Study on the tolerant function of soybean GmGST7 gene to acidic aluminum stress
-
摘要:目的
耐酸铝基因GsMYB7过表达转化大豆品种‘华春6号’后,从转基因株系的表达谱中获得目标基因GmGST7,该基因受酸铝胁迫诱导上调,且位于GsMYB7基因下游,进一步分析其耐酸铝功能,以期提高大豆酸铝耐受能力。
方法采用生物信息学方法分析GmGST7基因的碱基序列、蛋白结构域和构建系统进化树。通过烟草叶片瞬时转化法完成亚细胞定位。通过RT-qPCR分析该基因组织表达特异性。设计0、25、50、75 和 100 µmol/L 5个AlCl3浓度梯度,研究GmGST7对酸铝胁迫的响应。在50 µmol/L AlCl3处理下,设计0、4、8、12、16、24、36、48和72 h共9个时间梯度,对GmGST7的表达模式进行分析。过表达GmGST7基因遗传转化拟南芥,鉴定阳性植株,并对转基因株系进行耐酸铝表型验证、氧化水平测定、耐酸铝标志基因及下游基因的表达分析。
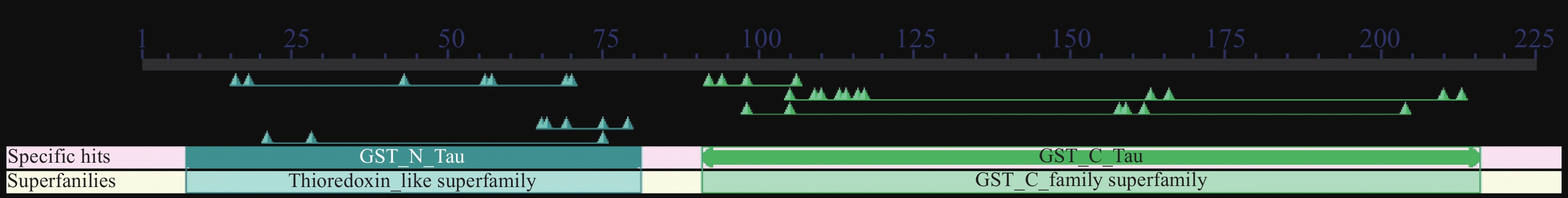
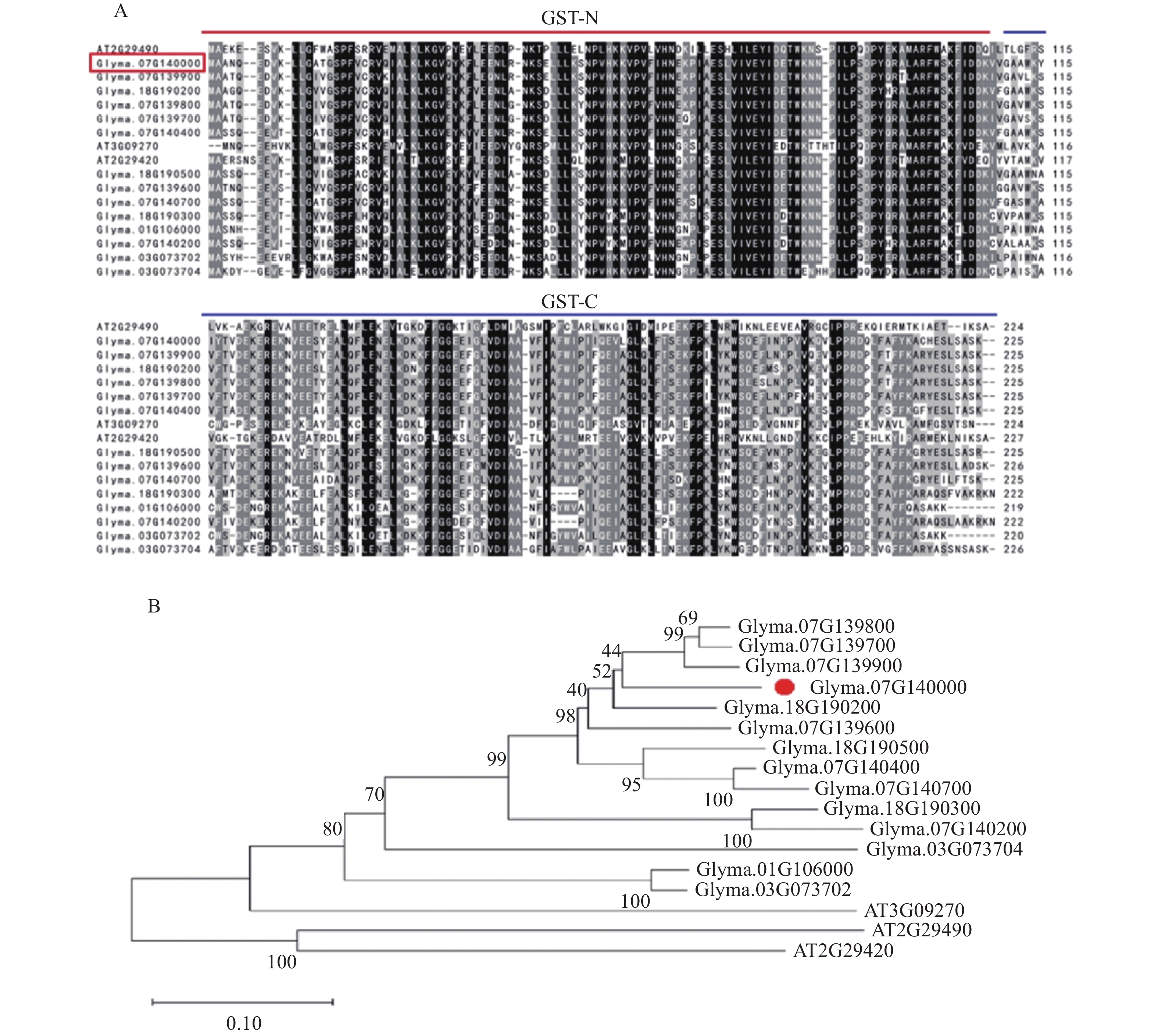
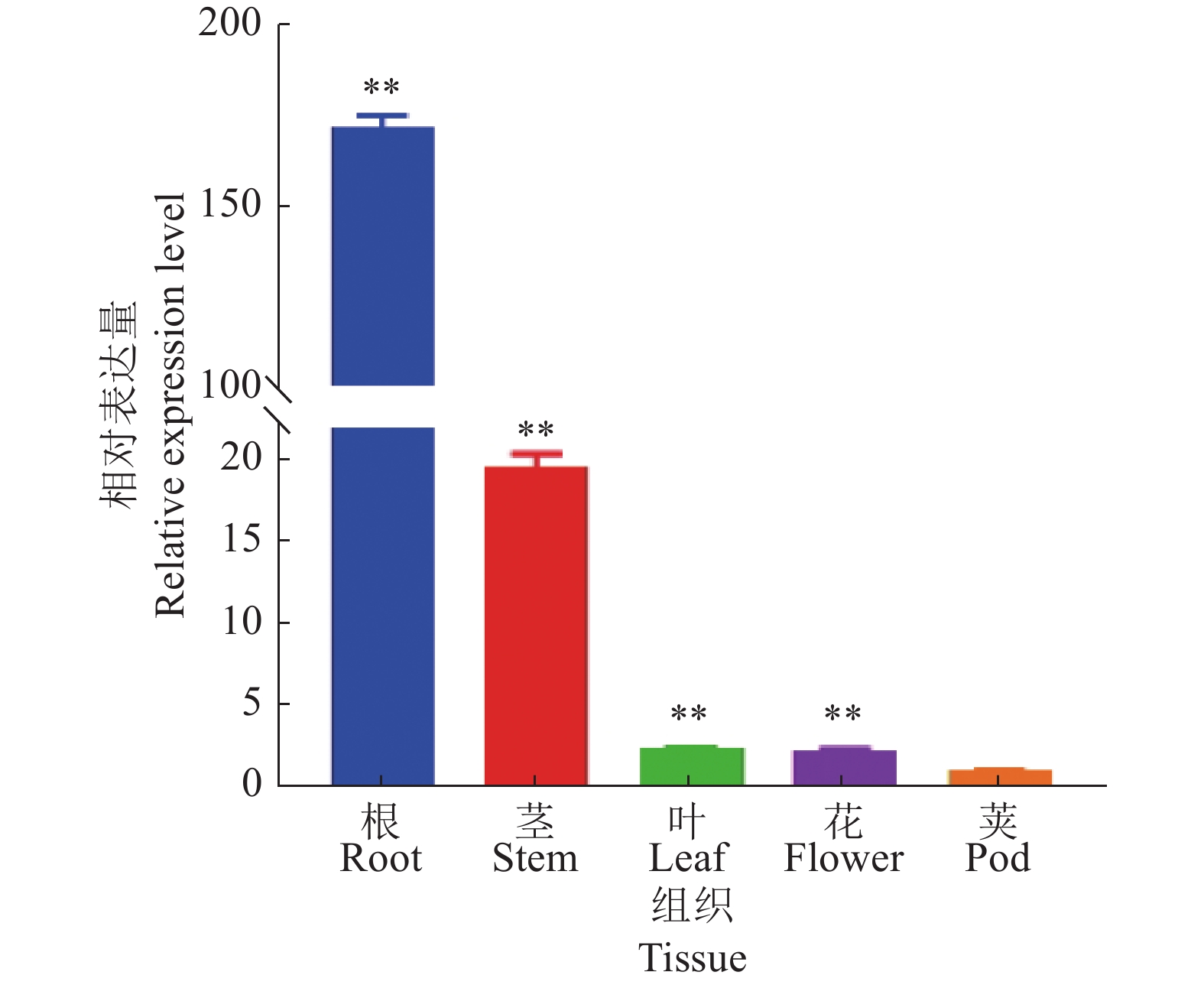
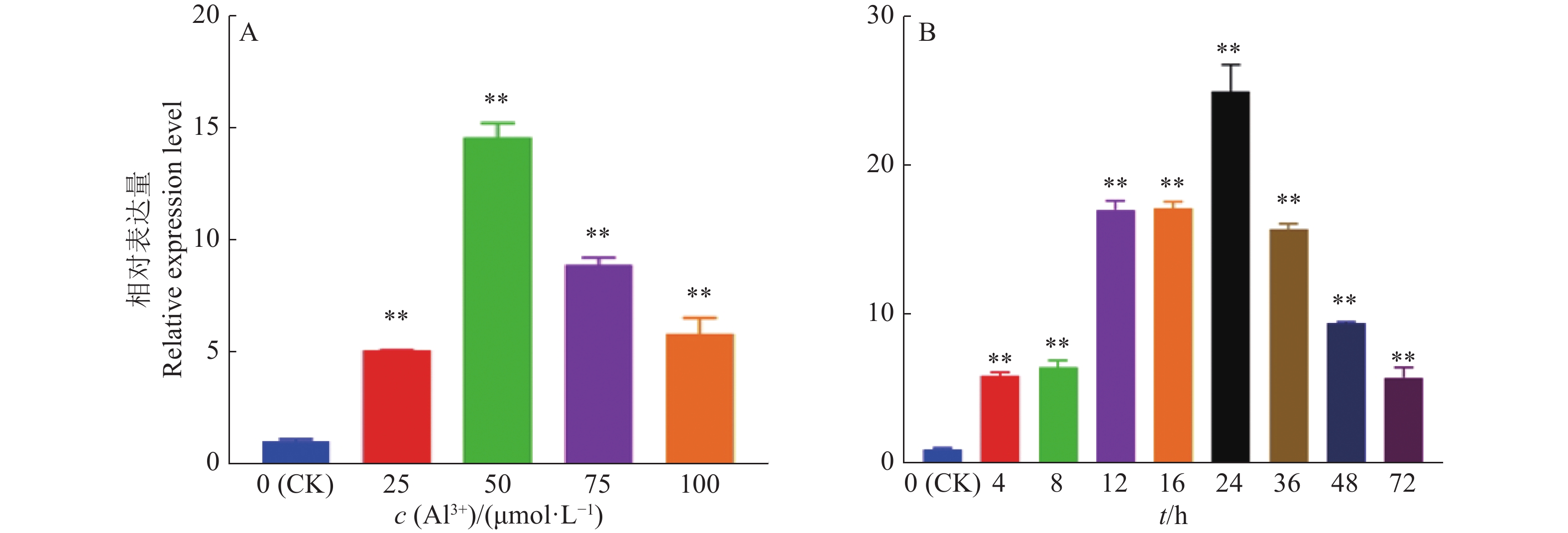
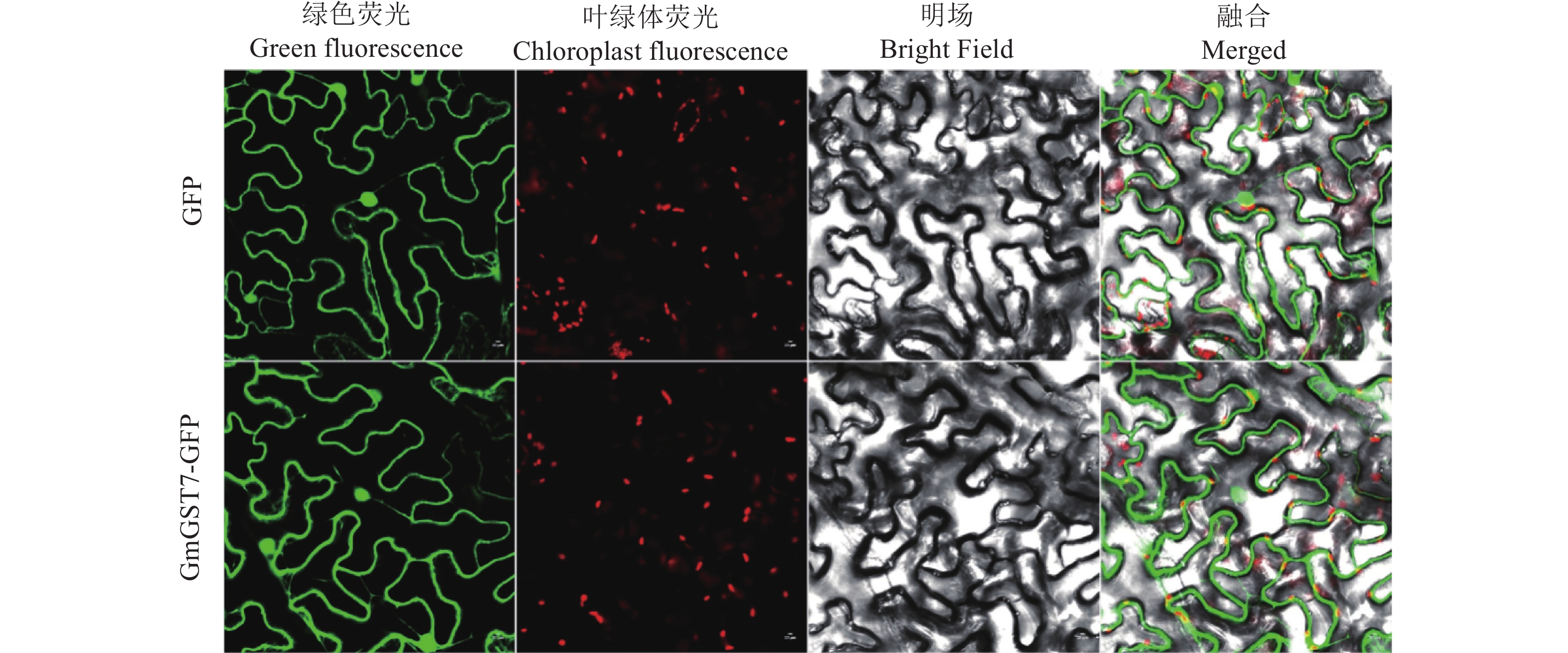
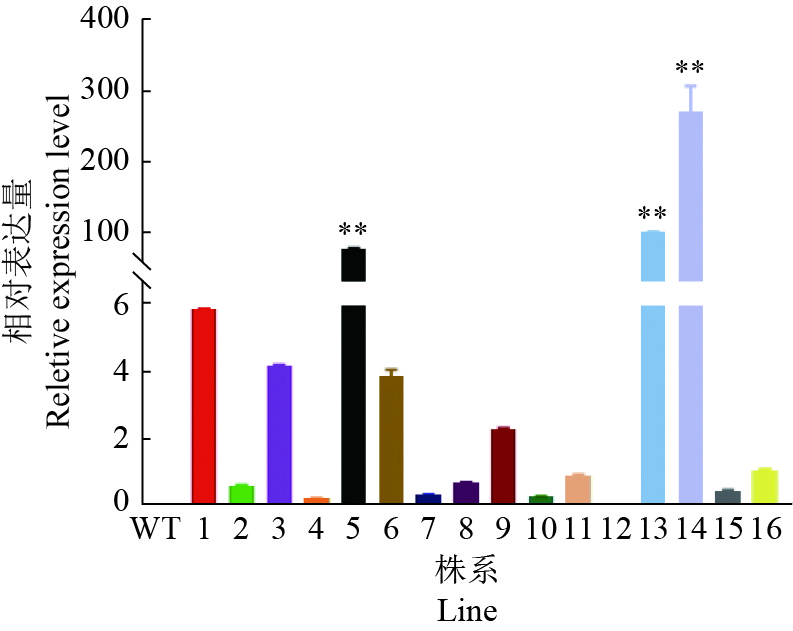
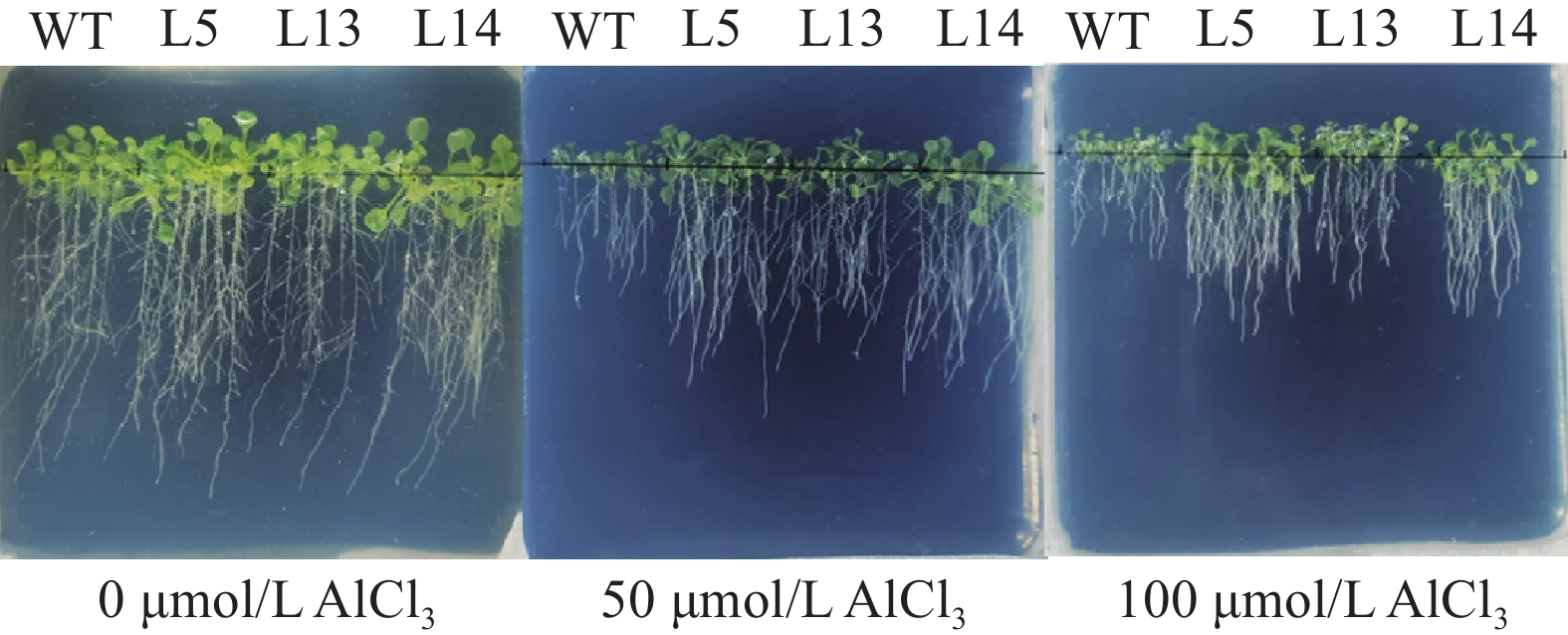
结果GmGST7基因位于大豆第7号染色体,序列全长为1 128 bp。该基因含有2个外显子和1个内含子,2个外显子分别编码GST高度保守的N端和不保守的C端;GmGST7基因编码226个氨基酸,编码的蛋白为大豆GST蛋白的tau类家族成员,定位于细胞质和细胞核中;GmGST7基因在大豆根、茎、叶、花和幼荚中均有表达,且在根中的表达量最高;GmGST7基因在50 µmol/L AlCl3处理24 h时表达最高;AlCl3处理后,野生型拟南芥相对根伸长显著低于转基因株系的,野生型拟南芥氧化水平高于转基因株系的,耐酸铝标志基因和下游基因的表达量在转基因株系中较高。
结论GmGST7基因属于大豆GST tau类家族成员,在细胞核和细胞质中行使功能,呈组成型表达模式,且在大豆根中表达最高,对酸铝胁迫响应显著;GmGST7过表达通过激活酸铝胁迫标志基因及其下游基因的表达提高拟南芥的酸铝耐受能力。
Abstract:ObjectiveThe GmGST7 gene was obtained from the gene expression profile of the GsMYB7 overexpressed lines of soybean ‘Huachun 6’ which was tolerant to acidic aluminum stress. GmGST7 lied downstream of the GsMYB7 gene, and was up-regulated by acidic aluminum stress. Its function of acidic aluminum resistance was further investigated to enhance the tolerance to aluminum stress in soybean.
MethodThe bioinformatics of the GmGST7 gene was analyzed using the base sequence, protein domain and phylogenetic tree. Subcellular localization of GmGST7 protein was accomplished by transient transformation in tobacco leaves. The tissue expression specificity of the GmGST7 gene was analyzed by RT-qPCR. Five AlCl3 concentration gradients of 0, 25, 50, 75 and 100 µmol/L were designed to study the response of GmGST7 to aluminum stress. Under the treatment of 50 µmol/L AlCl3, nine time gradients of 0, 4, 8, 12, 16, 24, 36, 48 and 72 h were designed to investigate the expression patterns of GmGST7. Arabidopsis (Col-0) was transformed by overexpression of GmGST7, positive plants were identified by molecular technology. The phenotype identification of Arabidopsis tolerant to acidic aluminum stress were performed with the oxidation level determination, the expression analysis of the genes response to aluminum stress and downstream genes of GmGST7.
ResultThe full-length sequence of GmGST7 located on chromosome 7 of soybean was 1 128 bp. The GmGST7 gene contained two exons and one intron which encodes a highly conserved N domain and a unconserved C domain of GST, respectively. GmGST7 encoded 226 amino acids. The GmGST7 protein was a tau member of the GST family in soybean, and localized in the nuclear and cytoplasm. GmGST7 was expressed in soybean root, stem, leaf, flower and young pod, and rich in root. The GmGST7 gene was up-regulated by AlCl3 with the highest relative expression under 50 µmol/L AlCl3 for 24 h. The relative root elongation of wild type was significantly lower than that of the transgenic lines, the oxidation level was higher, and the expression levels of acidic aluminum stress response genes and downstream genes were higher.
ConclusionThe GmGST7 gene is a tau member of the GST family in soybean, locates in the nucleus and cytoplasm. The GmGST7 gene holds a constitutive expression pattern, and is rich in soybean root. GmGST7 is significantly up-regulated by acidic aluminum stress. Overexpression of GmGST7 enhances the tolerance to aluminum stress in Arabidopsis by activating the expression of the marker genes response to acidic aluminum stress and its downstream genes.
-
Keywords:
- Soybean /
- GmGST7 /
- Acidic aluminum stress /
- Genetic transformation
-
由稻瘟病菌Magnaporthe oryzae引起的真菌病害稻瘟病是水稻重要病害之一,一般情况下发病会造成10%~20%的减产,流行年份可造成50%以上减产,严重影响水稻产量和品质,被称为“水稻癌症”[1-2]。根据稻瘟病发生的时期和部位,可分为苗瘟、叶瘟、节瘟和穗颈瘟等,其中穗颈瘟对水稻生产的危害最大,穗颈瘟一般发生在抽穗破口前,病菌侵入穗颈部并逐渐蔓延,阻断养分通道,严重威胁产量甚至造成绝收[3-4]。通过大量使用化学农药防治稻瘟病,不仅成本高,还易造成环境污染和农药残留超标,不利于生态环境和食品安全[5],且应用较广的三环唑、春雷霉素等均为预防性药剂,对防治窗口期短的穗颈瘟难以取得良好效果[6],聚合多个稻瘟病抗性基因、培育高抗品种是公认防控稻瘟病最经济、环保、有效的方式[7]。挖掘抗性基因是开展多基因聚合育种的重要前提,目前已鉴定的稻瘟病抗性基因超100个,已克隆的有39个[8],这为抗性基因的检测和利用提供了重要保障。抗病品种的选育依赖于抗病资源的发掘与利用。李刚等[9]鉴定了544份水稻种质资源的稻瘟病抗性水平及其携带的主效抗性基因,指出品种的抗性与其抗性基因种类密切相关,Pi5、Pita、Pi9和Pib对其研究所用的6个强致病小种抗性表现较好;朱业宝等[10]对156份外引水稻种质资源进行田间自然诱发鉴定和稻瘟病抗性基因检测,结果表明携带Pi9和Pi2的水稻资源的综合抗性较好,“Pi9+Pi5+Pik-m+Pia”、“Pi5+Pib+Pita+Pik-m+Pia”和“Pi2+Pi54+Pib+Pita+Pik-m+Pia”组合的稻瘟病综合抗性较好;王小秋等[11]分析了稻瘟病抗性基因在195个粳稻新品种/系中的分布情况,指出供试品种对穗颈瘟的抗性主要与Pia、Pi5和Pita显著相关,抗性基因组合“Pia+Pita”在江苏粳稻抗穗颈瘟育种中有重要应用价值;此外,周坤能等[12]、陈晴晴等[13]、王晓玲等[14]、潘争艳等[15]分别对153份安徽粳稻、252份长江中下游区试品种、82份江西粳籼骨干亲本、260份辽宁粳稻进行稻瘟病抗性基因检测及抗性鉴定,这些研究结果为各地抗性亲本的选择和品种的合理布局提供了良好的理论参考。但目前针对两广籼稻区优质常规稻育种骨干亲本的稻瘟病抗性基因及穗颈瘟抗性分析的研究鲜见报道。本研究以121份广西、广东历年审定的常规籼稻为材料,进行田间穗颈瘟抗性鉴定及Pi2、Pi5、Pi9、Pi33、Pi46、Pi54、Pib、Pid3、Pigm、Pit、Pita、Pia、Pik-m和Pik等14个稻瘟病抗性基因的分子鉴定,分析它们的基因型与抗性的关系,旨在为抗性基因聚合育种的亲本选择提供支持,为常规稻的合理布局提供理论参考。
1. 材料与方法
1.1 试验材料
试验材料为121份广西农业科学院水稻研究所优质常规稻育种研究室收集保存的常规籼稻,包括75份2000—2021年广西审定品种和46份1999—2020年广东审定品种。这些材料一方面曾作为华南稻区的主栽或主推品种,另一方面也是目前广西优质稻育种的骨干亲本。‘桂育6号’为广西水稻品种区试稻瘟病抗性鉴定所用的诱发材料。
1.2 试验方法
1.2.1 稻瘟病抗性基因检测
采用五引物扩增受阻突变体系(Penta-primer amplification refractory mutation system,PARMS) SNP分型技术进行Pi2、Pi5、Pi9、Pi33、Pi46、Pi54、Pib、Pid3、Pigm、Pit、Pita、Pia、Pik-m和Pik等14个稻瘟病抗性基因的检测,DNA提取采用常规的CTAB法,PCR反应体系10 μL,35个循环,分子特异性标记引物由武汉市景肽生物科技有限公司提供。PCR完成后,以FAM和HEX作为报告荧光,ROX作为参比荧光,用TECAN infinite F200酶标仪读取荧光信号,解析转换荧光信号得到清晰直观的分型图,并根据颜色不同,输出基因型结果[5, 16]。具体检测委托武汉市景肽生物科技有限公司进行。
1.2.2 稻瘟病田间抗性鉴定
自然诱发鉴定于2022年早稻在广西壮族自治区来宾市金秀瑶族自治县罗香乡琼伍村旱田屯(110.09°E、24.39°N)进行,该试验地处山谷丘陵,海拔270 m,雾大露重,稻分蘖盛期日平均气温为20~25 ℃,相对湿度为90%以上,具有诱发稻瘟病的有利环境条件。每份材料种植1个小区,每小区10行,每行4株,小区两边各插1列诱发材料,株行距20.0 cm×13.0 cm,小区随机排列,重复3次。除不进行稻瘟病防治外,田间其他管理同一般水田。于黄熟初期按《水稻品种试验稻瘟病抗性鉴定与评价技术规程(NY/T2646—2014)》[17]的方法调查穗颈瘟,并按该标准中的“水稻穗颈瘟单穗损失率0~9 级分级标准”和“水稻穗颈瘟发病率群体抗性分级标准”对材料进行穗颈瘟抗性分级,以3次重复中感病最严重的程度作为鉴定结果。
1.3 统计分析
采用办公软件WPS对研究中获得的各类数据进行整理和作图。以DPS v9.01进行统计分析,采用LSD法检测差异显著性。
采用SPSS statistics 26进行逻辑回归分析[11],将供试材料田间穗颈瘟抗级≤3的定义为1, > 3的定义为0,携带某抗性基因定义为1,不携带该抗性基因定义为0,得到数据表后对单个基因进行逻辑回归分析及卡方检测,获得各基因与抗性间的回归系数(B)、显著性检验的P值、优势比(Odds ratio,OR)和95% 置信区间(Confidence interval,CI)。
2. 结果与分析
2.1 稻瘟病抗性基因检测
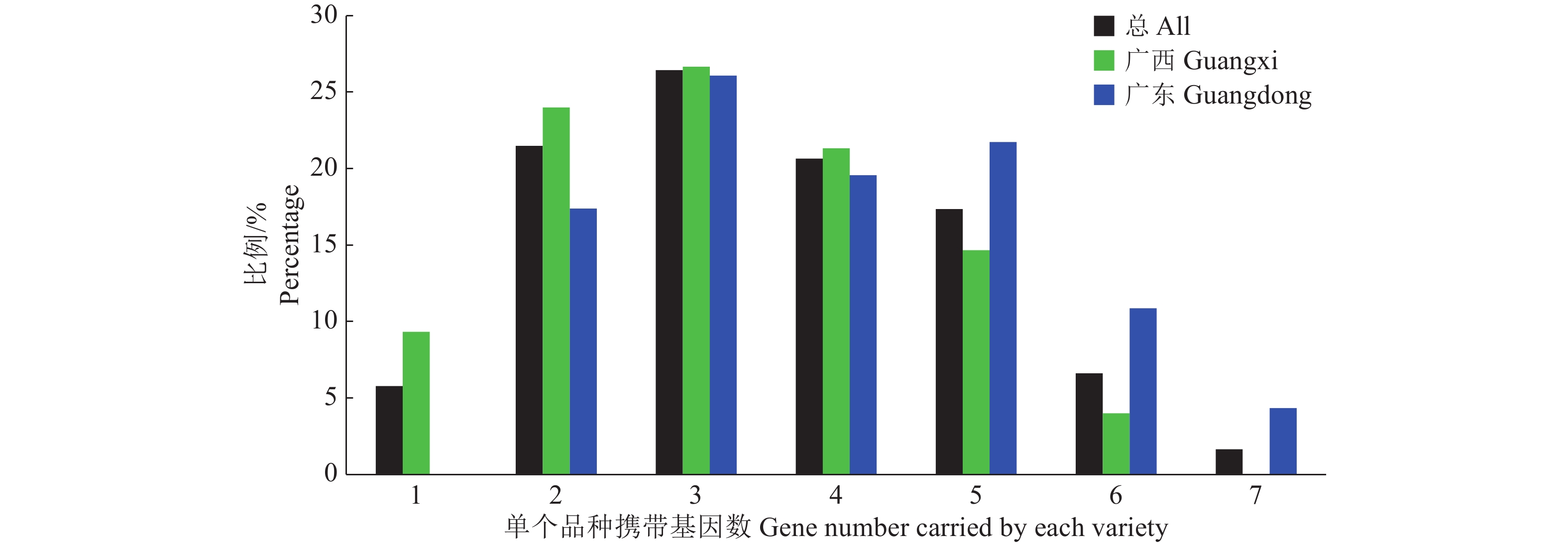
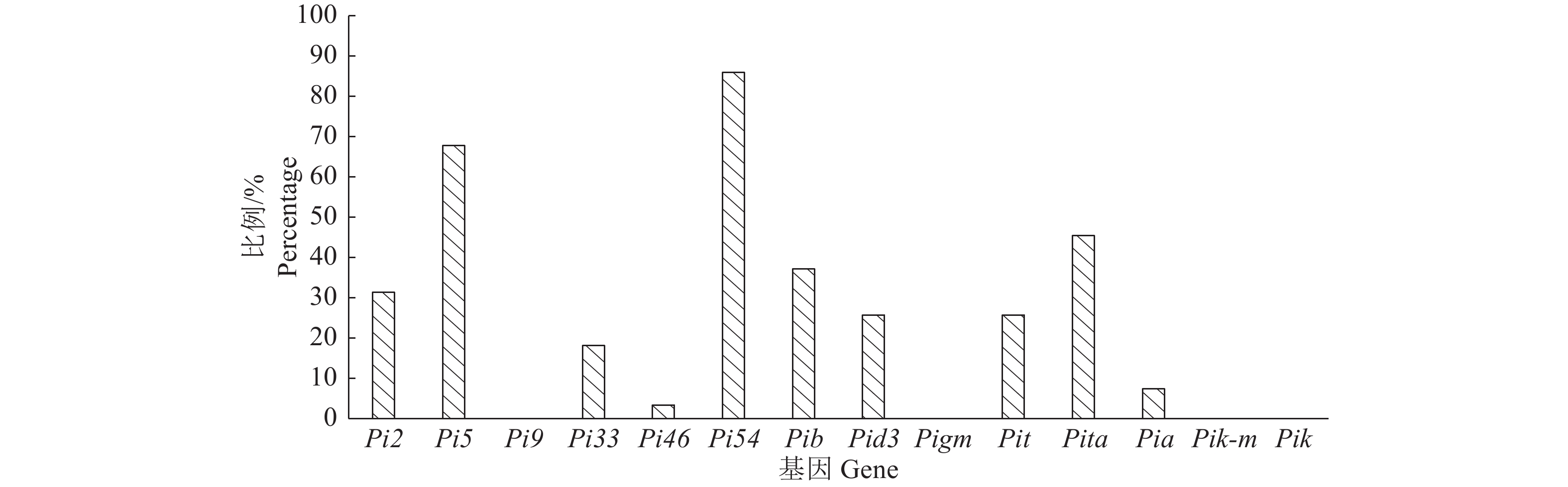
利用PARMS SNP分型技术对稻瘟病抗性基因Pi2、Pi5、Pi9、Pi33、Pi46、Pi54、Pib、Pid3、Pigm、Pit、Pita、Pia、Pik-m和Pik的检测结果(图1)表明,所有供试品种均不携带Pi9、Pigm、Pik-m和Pik,其余10个抗性基因则有不同程度的分布;供试品种中,Pi46和Pia的检出率较低,分别为3.3%和7.4%,Pi54检出率高达86.0%,Pi5检出率为67.8%,其他基因检出率在18%~46%之间;就品种而言(图2),携带了2~5个基因的品种占总数的86.0%,携带1个基因的品种有7份,均为广西品种,携带7个基因的品种有2份,均为广东品种。以上结果表明,供试品种在携带已知稻瘟病抗性基因上存在明显差异,Pi54和Pi5的应用较广,Pi9、Pigm、Pik-m和Pik在供试品种中尚没有被利用。
2.2 稻瘟病抗性基因在不同类型供试品种中的应用情况
为分析不同地域环境、审定年份的品种间携带的抗性基因是否存在差异,比较除供试品种都不携带的Pi9、Pigm、Pik-m和Pik外其他抗性基因在不同类型供试品种中的分布,结果(表1)显示,2011年后的广西品种较2011年前相比,Pi2、Pi5、Pid3和Pit出现频率分别提高22.7、22.8、18.2和20.3个百分点,特别是Pi5在2011年后出现频率达84.1%,而Pi46和Pita出现频率分别降低12.9和15.2个百分点;2011年后的广东品种较2011年前相比,Pi2、Pi33、Pib和Pid3出现频率分别提高42.7、16.2、20.4和35.4个百分点,特别是Pi2在2011年后出现频率达85.0%,而Pi5和Pita出现频率分别降低29.2和22.7个百分点;综合比较广西和广东品种间抗性基因分布频率,Pi2和Pid3在广西品种中出现频率比广东品种分别低47.6和39.3个百分点,而Pi5在广西品种中出现频率高出广东品种18.2个百分点。以上结果表明,多数抗性基因在不同类型品种中应用基本相近,广东常规稻选育中主要是对抗性基因Pi2、Pi5、Pi54、Pid3和Pita的利用,广西常规稻选育中主要是对抗性基因Pi5、Pi54和Pita的利用,而2011年后广西的常规稻选育中对Pi2和Pid3的利用也明显提升。
表 1 不同类型供试品种间抗性基因分布频率Table 1. Distribution frequency of resistance genes in different types of tested varieties% 品种类型
Type of varietyPi2 Pi5 Pi33 Pi46 Pi54 Pib Pid3 Pit Pita Pia 广西品种
Guangxi varieties13.3 74.7 22.7 5.3 81.3 33.3 10.7 28.0 44.0 5.3 广东品种
Guangdong varieties60.9 56.5 10.9 0 84.3 43.5 50.0 21.7 47.8 10.9 2011年前广西品种
Guangxi varieties before 20110 61.3 25.8 12.9 80.6 32.3 0 16.1 51.6 6.5 2011年前广东品种
Guangdong varieties before 201142.3 69.2 3.8 0.0 92.3 34.6 34.6 23.1 57.7 7.7 2011年后广西品种
Guangxi varieties after 201122.7 84.1 20.5 0.0 81.8 34.1 18.2 36.4 36.4 4.5 2011年后广东品种
Guangdong varieties after 201185.0 40.0 20.0 0.0 95.0 55.0 70.0 20.0 35.0 15.0 2.3 供试品种穗颈瘟抗性分析
穗颈瘟田间抗性调查结果(表2)显示,供试品种中没有高抗穗颈瘟的品种,中抗+抗的品种有25份,占比20.6%,感+高感的比例高达到51.3%;广西的供试品种没有达高抗或抗水平的,中抗品种也仅占比10.7%,感+高感的比例高达到64.0%;广东的供试品种中抗+抗的品种有17份,占比37.0%,感+高感的品种有14份,占比30.4%。以上结果表明,供试品种的穗颈瘟抗性普遍较弱,相比之下,广东供试品种的穗颈瘟抗性总体明显好于广西供试品种。
表 2 供试品种穗颈瘟抗级分布频率Table 2. Distribution frequency of resistance level to panicle neck blast in tested varieties% 品种类型
Type of variety0
(高抗 Highly resistant)1
(抗 Resistant)3
(中抗 Medium resistrant)5
(中感 Medium susceptible)7
(感 Susceptible)9
(高感 Highly susceptible)总供试品种
All tested vararieties0 4.9 15.7 28.1 18.2 33.1 广西品种
Guangxi varieties0 0 10.7 25.3 18.7 45.3 广东品种
Guangdong varieties0 13.1 23.9 32.6 17.4 13.0 2.4 携带的抗性基因数量与穗颈瘟抗性相关分析
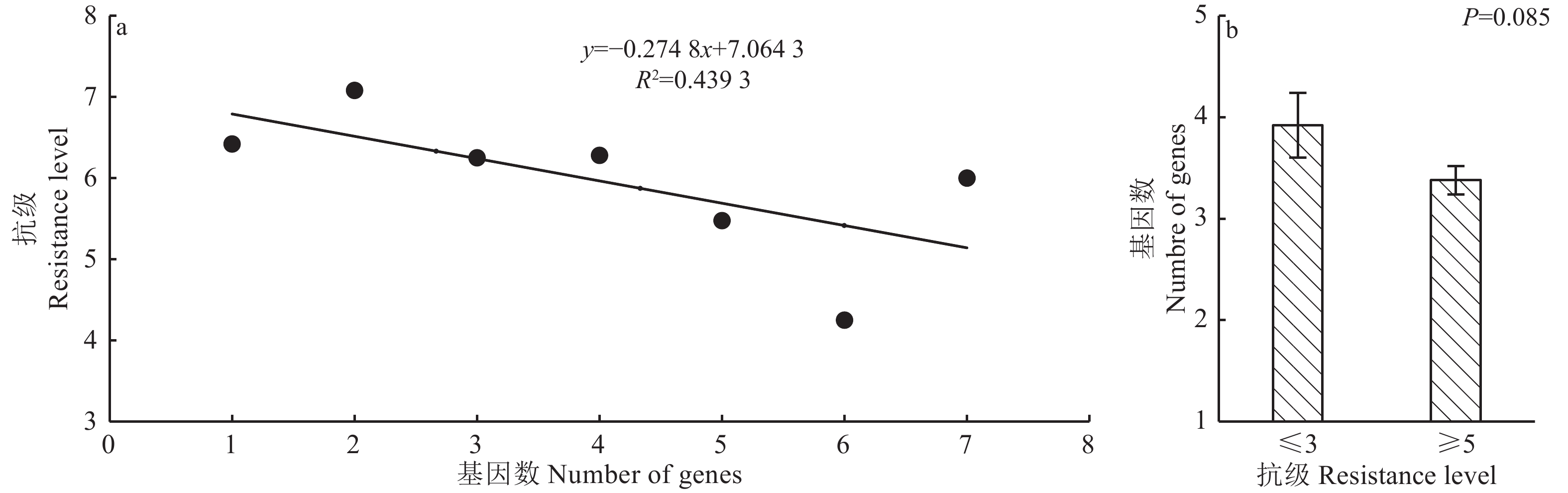
供试品种携带的抗性基因数量与抗级间的相关分析显示(图3a),二者间的决定系数R2=0.4393,未达统计显著水平(P=0.105),表明抗性基因数量与穗颈瘟抗性间的相关性不显著。进一步比较抗的(抗级≤3)和感的(抗级≥5)供试品种间携带的抗性基因数差异(图3b),发现2类品种携带的平均抗性基因数分别为3.92和3.38,差异不显著(P=0.085)。同时在供试品种中也发现携带的抗性基因数量少却能达到中抗及以上水平的品种,及携带抗性基因数量多却表现中感以下的品种。因此对品种的穗颈瘟抗性改良,除简单的抗性基因聚合外,还应考虑不同遗传背景下抗性基因的强弱及组合效应。
2.5 稻瘟病抗性基因对穗颈瘟抗性贡献分析
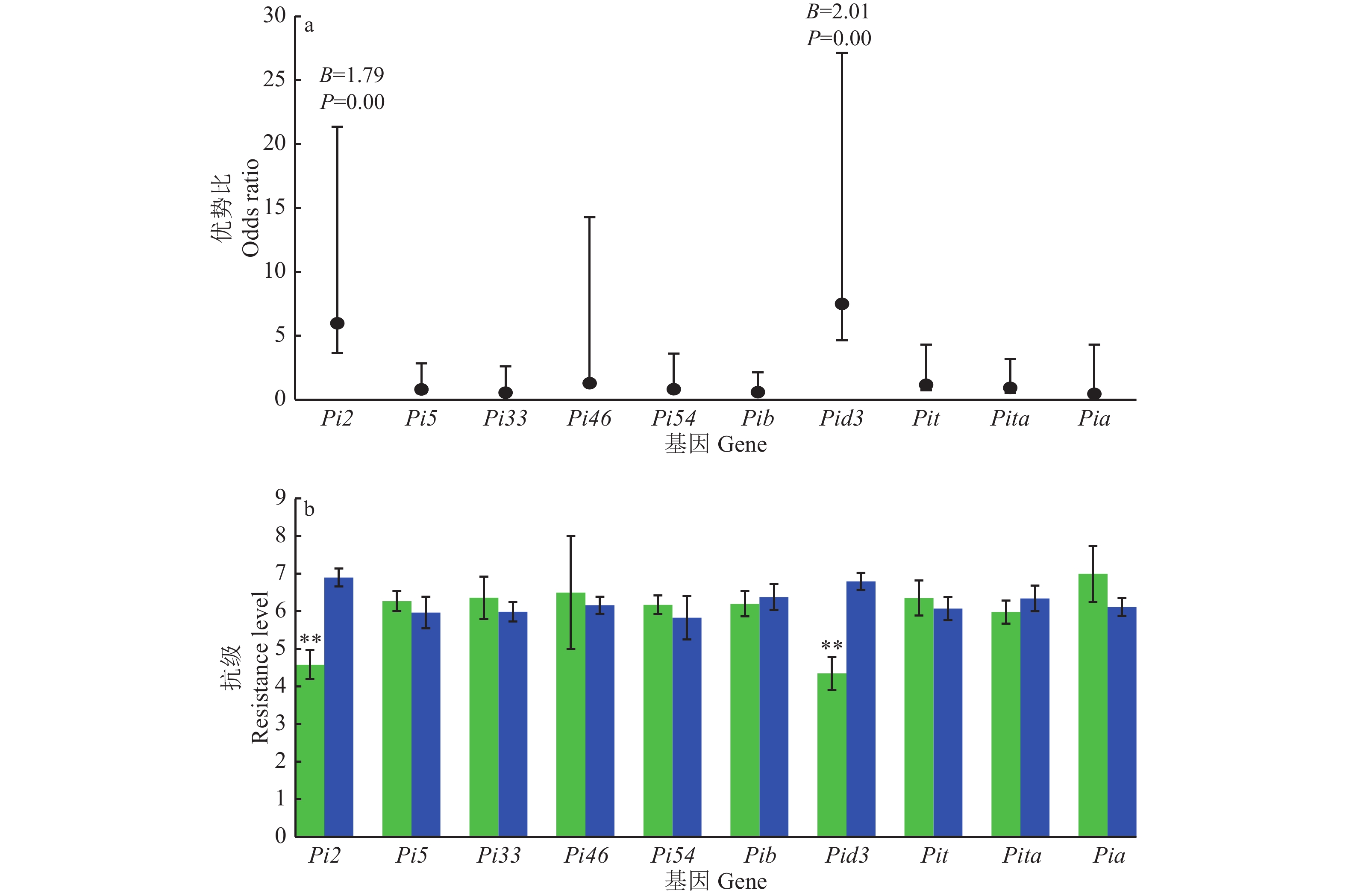
采用逻辑回归法分析各基因对穗颈瘟抗性的贡献,结果见图4a,Pi2与穗颈瘟抗性间的回归系数(B)为1.79,达到极显著(P=0.00),Pid3与穗颈瘟抗性间的B为2.01,达到极显著(P=0.00),表明在单基因水平上Pi2和Pid3对抗性有显著贡献;Pi2和Pid3的优势比(OR)分别为5.98和7.50,表明携带Pi2、Pid3对抗性的贡献是不携带Pi2、Pid3的5.98与7.50倍。进一步分析(图4b)显示,携带Pi2(Pi2+)的品种平均抗级为4.58,不携带Pi2(Pi2−)的品种平均抗级为6.90,差异极显著,携带Pid3(Pid3+)的品种平均抗级为4.35,不携带Pid3(Pid3−)的品种平均抗级为6.80,差异极显著,而其他基因的携带与否,造成的抗级差异均不显著。
![]() 图 4 单个抗性基因对穗颈瘟抗性贡献的逻辑回归分析(a)及平均抗级(b)图a中黑点上下黑线分别为各优势比的95%置信区间;图b中绿色柱表示携带某基因的平均抗级,蓝色柱表示不携带某基因的平均抗级,“**”表示与不携带该基因的材料差异显著(P<0.01, t检验)Figure 4. Logistic regression analysis of contribution to resistance (a) and average resistance (b) of single gene to panicle neck blastIn figure a, the black lines above and below each black dot represent the 95% confidence intervals for each odds ratio; In figure b, the green column represents the average resistance of carrying a certain gene, the blue column represents the average resistance level of not carrying a certain gene , and “**” indicates significant difference from the material not carrying the gene (P<0.01, t test)
图 4 单个抗性基因对穗颈瘟抗性贡献的逻辑回归分析(a)及平均抗级(b)图a中黑点上下黑线分别为各优势比的95%置信区间;图b中绿色柱表示携带某基因的平均抗级,蓝色柱表示不携带某基因的平均抗级,“**”表示与不携带该基因的材料差异显著(P<0.01, t检验)Figure 4. Logistic regression analysis of contribution to resistance (a) and average resistance (b) of single gene to panicle neck blastIn figure a, the black lines above and below each black dot represent the 95% confidence intervals for each odds ratio; In figure b, the green column represents the average resistance of carrying a certain gene, the blue column represents the average resistance level of not carrying a certain gene , and “**” indicates significant difference from the material not carrying the gene (P<0.01, t test)2.6 稻瘟病抗性基因组合效应分析
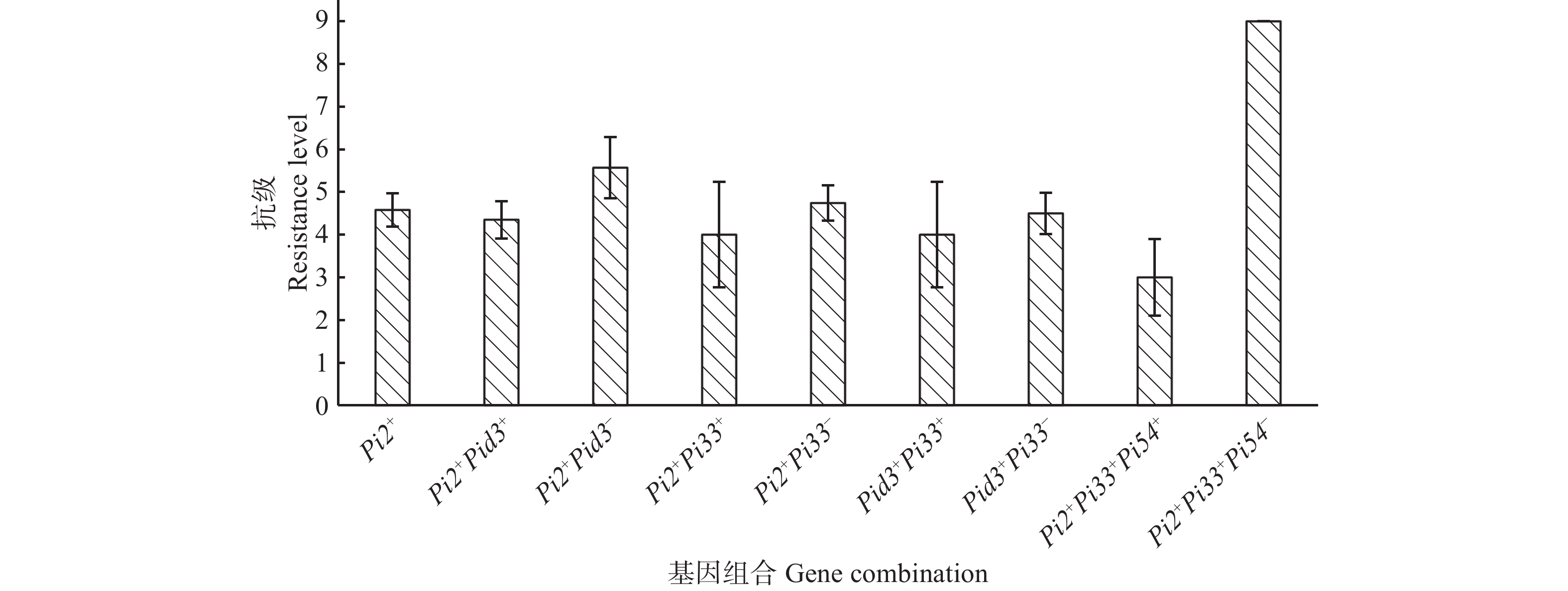
分析抗性基因间的聚合叠加效应,结果(图5)显示,Pi2+Pid3+平均抗级4.35,Pi2+Pid3−的平均抗级5.57,Pi2+的平均抗级4.58,表明聚合Pi2和Pid3能增强抗病效应;Pi2+Pi33+平均抗级低于Pi2+Pi33−和Pi2+的平均抗级,Pid3+Pi33+平均抗级低于Pid3+Pi33−的平均抗级,表明在携带Pi2和Pid3的材料中聚合Pi33能一定程度上增强抗病效应。此外,5个Pi2+Pi33+Pi54+的供试品种平均抗级3.0,田间穗颈瘟鉴定无感~高感的情况,1个Pi2+Pi33+Pi54−的供试品种抗级9.0,表明Pi54也可能使Pi2+Pi33+的抗病效应增强。
3. 讨论与结论
稻瘟病生理小种复杂多变,不同的水稻种植区域优势小种构成往往不同,稻瘟病抗性基因在面对不同的生理小种时表现的抗性也不尽相同,培育广谱、持久抗性的水稻品种是防控稻瘟病的重要措施。而抗性基因鉴定及田间抗性分析,能较快掌握目标基因的分布状态和潜在的育种价值,是水稻育种资源鉴定的重要基础。
陆展华等[18]对广东省主栽品种的稻瘟病抗性基因及田间抗性鉴定表明,Pi2的检出率为17.6%,且抗性贡献显著,Pib和Pita的检出率虽高,但抗性贡献低;Pi9作为广谱抗性基因,对大多数国家的生理小种均表现出较好的抗性[19-20],但Pi9在国内育种上的应用差异较大,在东北稻区粳稻品种中检出率高达45.8%[21],而在广东籼稻主栽品种中未鉴定到[22],Tian等[23]研究也认为Pi9还未广泛应用于我国籼稻品种。本研究结果发现,来源于两广地区的供试籼型常规稻品种均不携带Pi9,Pi2的检出率为31.4%,逻辑回归分析表明Pi2对穗颈瘟抗性贡献极显著,Pib和Pita的检出率分别为37.2%和45.5%,均对穗颈瘟抗性贡献不显著,本研究与上述研究结论是一致的,说明Pib和Pita对两广籼稻区的生产应用价值降低,应持续加强对Pi2的应用,并挖掘Pi9在两广籼稻区抗病品种选育及生产上的应用潜力。主效稻瘟病抗性基因Pik及其等位基因Pik-m对国内多个生理小种都有良好的抗性[24],Pik-m和Pik对广东、湖南、四川、江苏等稻瘟菌株的抗性频率达到90%以上[25],广谱抗性基因Pigm是一个与Pi2等位、抗谱差异显著的基因,对稻瘟病抗性改良有重要应用价值[26],本研究结果发现供试品种均不携带Pik-m、Pik和Pigm,暗示着两广籼稻区抗病品种选育及生产可尝试对Pik-m、Pik和Pigm的利用。Pid3是从‘谷梅2号’中克隆的一个广谱稻瘟病抗性基因,在抗病品种改良中具有重要的利用价值[27],本研究结果表明,Pid3对穗颈瘟抗性贡献显著,供试品种中Pid3的平均检出率为25.6%,2011年前后广西供试品种中Pid3的检出率从0提高到18.2%,2011年前后广东供试品种中Pid3的检出率从34.6%提高到70.0%,说明对Pid3在抗病育种中的重视和利用逐渐加强。
本研究结果发现,对穗颈瘟抗性贡献显著的Pi2和Pid3在广东品种中的应用明显高于广西品种,这是广东品种田间穗颈瘟抗性表现明显好于广西品种的重要原因之一。2011年以前广西品种对Pi2和Pid3的利用为0,2011年之后广西常规稻品种选育中加强了对这2个基因的利用,这也是广西常规稻品种稻瘟病抗性提升的重要原因之一。
利用分子标记辅助选择是水稻新品种选育的高效手段,聚合多个稻瘟病抗性基因是改良品种抗性的常用方法,陈晴晴等[13]研究表明,聚合3个抗性基因的穗颈瘟抗病率比1个基因时上升54.3%,但更多的研究认为,携带的抗性基因数与稻瘟病抗性之间并不呈显著的负相关[11-12],抗性基因的聚合并非简单的抗谱叠加,应选择抗性好、抗谱互补的基因进行聚合,不仅能拓宽抗谱,还能提高对一些生理小种的抗性[28]。与前人研究结果一致,本研究结果表明,携带的抗性基因数量与穗颈瘟抗性间相关性不显著,对穗颈瘟的抗性改良,除简单的抗性基因聚合外,还应该充分考虑不同遗传背景下抗性基因的强弱及基因间的互作效应,而本研究对稻瘟病抗性基因组合效应分析发现,Pi2+Pid3+、Pi2+Pi33+和Pid3+Pi33+组合的穗颈瘟抗性表现较好。此外本研究中6个Pi2+Pi33+Pi54+的供试品种平均抗级3.0,唯一一个Pi2+Pi33+Pi54−的供试品种抗级9.0,是否是Pi54的存在增强了Pi2+Pi33+的抗性,还需更多的样本支撑。
-
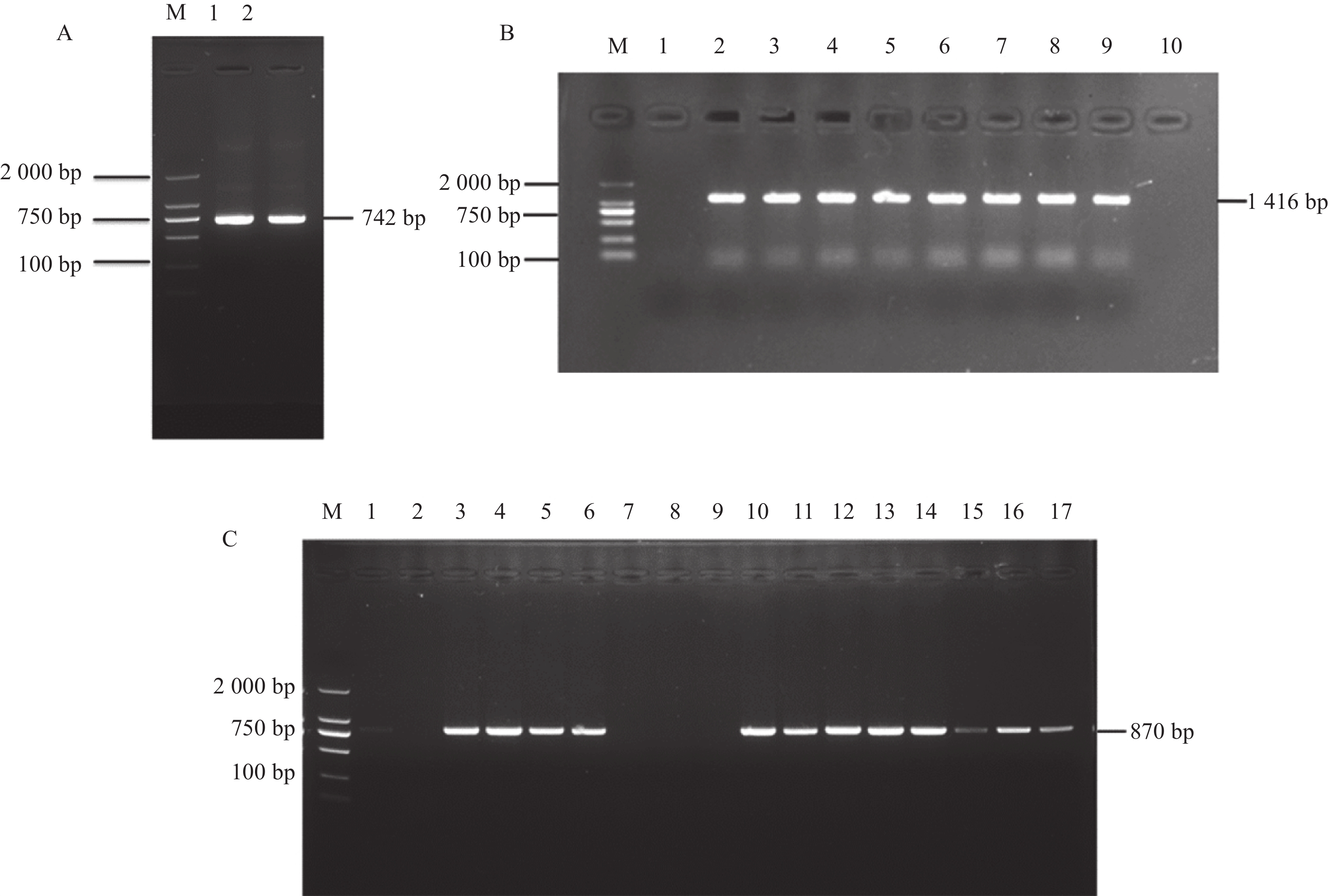
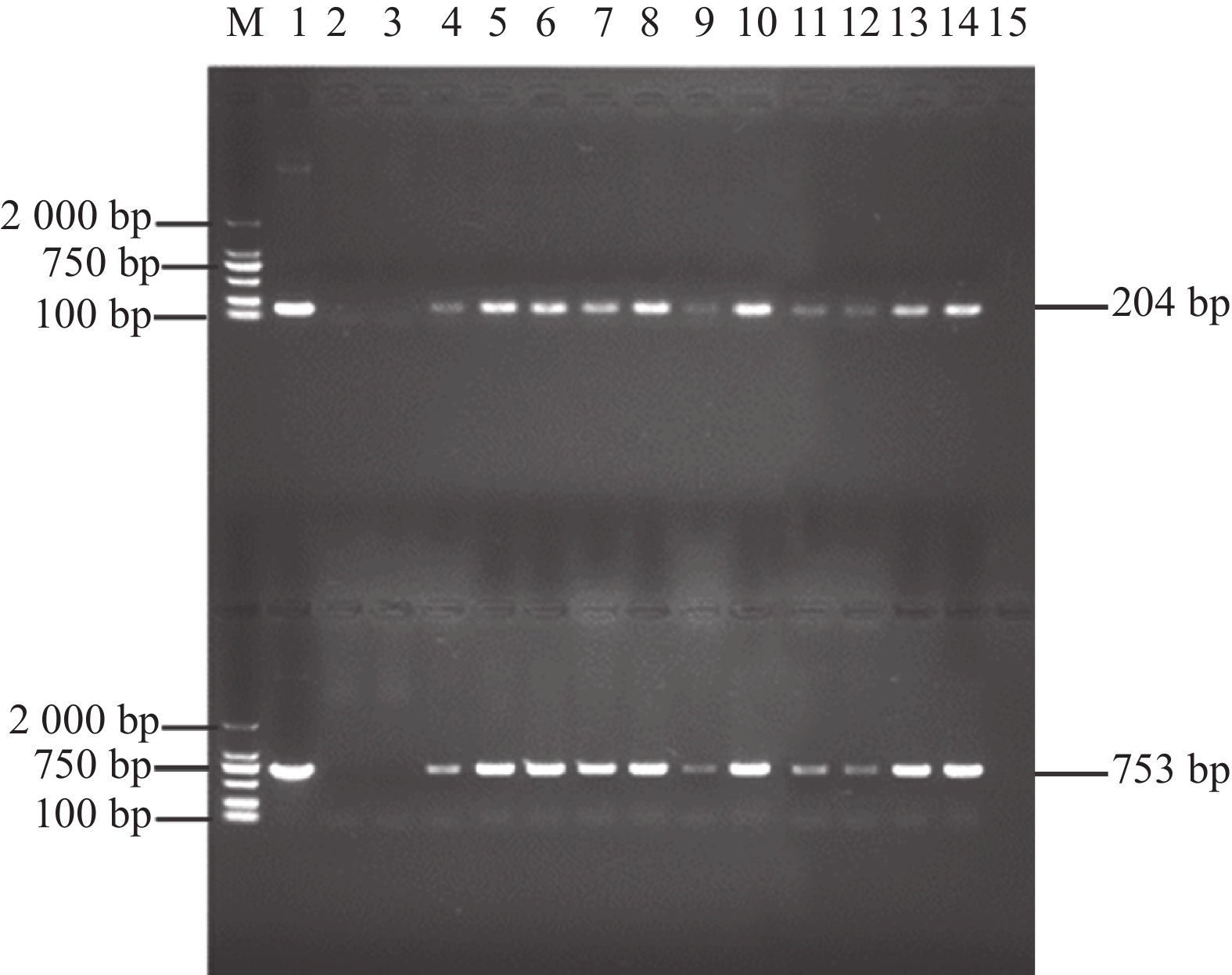
图 6 GmGST7基因克隆及载体构建
A:GmGST7基因cDNA序列克隆,根据引物位置克隆大小为742 bp;B:连亚细胞定位载体后,根据载体引物位置PCR产物大小为1 416 bp;C:连接过表达载体后,根据载体引物位置PCR产物大小为870 bp
Figure 6. Cloning and vector construction of the GmGST7 gene
A: Cloning of GmGST7 gene cDNA sequence, according to the location of the primer, cloning size is 742 bp; B: After connecting the subcellular localization vector, according to the location of the carrier primer, the size of the PCR product is 1 416 bp; C: After connecting the overexpressed vector, according to the location of the vector primer, the size of the PCR product was 870 bp
表 1 引物序列
Table 1 Primer sequence
引物名称
Primer name引物序列(5′→3′)
Primer sequencepTF101-GmGST7-F gagaacacgggggactctagaATGGCTGCTAATCAGGAAGATGTG pTF101-GmGST7-R cgatcggggaaattcgagctcTTTTGAAGCAGAAAGACTTTCATGG Super1300-GmGST7-F acgggggactcttgaccatggCTATGGCTGCTAATCAGGAAGATGTG Super1300-GmGST7-R aagttcttctcctttactagtTTTTGAAGCAGAAAGACTTTCATGG GmGST7-F GTCCTGATTCCCGGCTCAAT GmGST7-R AACTCACAAATGAGAGACCAGT RT-GmGST7-F TCAACCACCCTGTTGTCAAAC RT-GmGST7-R AAGACTTTCATGGCAGGCTTTGT 表 2 荧光定量引物序列
Table 2 qPCR primer sequence
基因
Gene引物序列(5′→3′)
Primer sequenceAtALMT F: TCCCATGGGTAAAGACAAAG
R: ATAGTCTGCTTTCTGCCAAAAtMATE F: CATTCGAATCCATCGAGATT
R: CGAATGTTGCACTCTGTTTTAtALS3 F: AGCTTCGAGATGACATCAAA
R: ACGGTTTTGCAGCTATCTAAAtWAK1 F: TGGCCGCTGATATTACAAAT
R: CAGATTGGCTACTGGTTAGTAt1G78660 F: CAGGTTTGAGTGTATCGGTG
R: CATCTGATTCTTCTGCCCAAAt1G78670 F: TCCTCTGAGATGTGGAGATT
R: TAGTTGAGGTTTGGATCAGCAt1G78680 F: AAAATGGTGGATTTTGCAGG
R: ATAGGCTGACGTTCAAAGTTAt4G33090 F: TGGATCAGTTCAAAGGTGAG
R: GACTATGTCGAGATCGATGG -
[1] VON UEXKÜLL H R, MUTERT E. Global extent, development and economic impact of acid soils[J]. Plant and Soil, 1995, 171(1): 1-15. doi: 10.1007/BF00009558
[2] KINRAIDE T B, PARKER D R. Cation amelioration of aluminum toxicity in wheat[J]. Plant Physiology, 1987, 83(3): 546-551. doi: 10.1104/pp.83.3.546
[3] KINRAIDE T B. Assessing the rhizotoxicity of the aluminate ion, Al(OH)4−[J]. Plant Physiology, 1990, 93(4): 1620-1625. doi: 10.1104/pp.93.4.1620
[4] DELHAIZE E, RYAN P R, RANDALL P J. Aluminum tolerance in wheat (Triticum aestivum L.) II: Aluminum-stimulated excretion of malic acid from root apices)[J]. Plant Physiology, 1993, 103(3): 695-702. doi: 10.1104/pp.103.3.695
[5] SIVAGURU M, BALUSKA F, VOLKMANN D, et al. Impacts of aluminum on the cytoskeleton of the maize root apex. short-term effects on the distal part of the transition zone[J]. Plant Physiology, 1999, 119(3): 1073-1082. doi: 10.1104/pp.119.3.1073
[6] KOLLMEIER M, FELLE H H, HORST W J. Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum?[J]. Plant Physiology, 2000, 122(3): 945-956. doi: 10.1104/pp.122.3.945
[7] VAZQUEZ M D, POSCHENRIEDER C, et al. Change in apoplastic aluminum during the initial growth response to aluminum by roots of a tolerant maize variety[J]. Plant Physiology, 1999, 119(2): 435-444. doi: 10.1104/pp.119.2.435
[8] SILVA I R, SMYTH T J, MOXLEY D F, et al. Aluminum accumulation at nuclei of cells in the root tip. Fluorescence detection using lumogallion and confocal laser scanning microscopy[J]. Plant Physiology, 2000, 123(2): 543-552. doi: 10.1104/pp.123.2.543
[9] TAYLOR G J, MCDONALD-STEPHENS J L, HUNTER D B, et al. Direct measurement of aluminum uptake and distribution in single cells of Chara corallina[J]. Plant Physiology, 2000, 123(3): 987-996. doi: 10.1104/pp.123.3.987
[10] FRANTZIOS G, GALATIS B, APOSTOLAKOS P. Aluminium causes variable responses in actin filament cytoskeleton of the root tip cells of Triticum turgidum[J]. Protoplasma, 2005, 225(3/4): 129-140.
[11] ČIAMPOROVÁ M. Morphological and structural responses of plant roots to aluminium at organ, tissue, and cellular levels[J]. Biologia Plantarum, 2002, 45(2): 161-171. doi: 10.1023/A:1015159601881
[12] JONES D L, KOCHIAN L V. Aluminum inhibition of the inositol 1, 4, 5-trisphosphate signal transduction pathway in wheat roots: A role in aluminum toxicity?[J]. The Plant Cell, 1995: 1913-1922.
[13] BARCELÓ J, POSCHENRIEDER C. Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: A review[J]. Environmental and Experimental Botany, 2002, 48(1): 75-92. doi: 10.1016/S0098-8472(02)00013-8
[14] KOCHIAN L V, PIÑEROS M A, HOEKENGA O A. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity[J]. Plant and Soil, 2005, 274(1/2): 175-195. doi: 10.1007/s11104-004-1158-7
[15] WANG H J. GsMYB7 encoding a R2R3-type MYB transcription factor enhances the tolerance to aluminum stress in soybean (Glycine max L.) [J]. BMC Genomics, 2022. 23(1): 529. doi: 10.1186/s12864-022-08744-w.
[16] SHEEHAN D, MEADE G, FOLEY V M, et al. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily[J]. The Biochemical Journal, 2001, 360(Pt1): 1-16.
[17] SAADAT M, MOHABATKAR H. Polymorphisms of glutathione S-transferases M1 and T1 do not account for inter individual differences for smoking behavior[J]. Pharmacology Biochemistry and Behavior, 2004, 77(4): 793-795. doi: 10.1016/j.pbb.2004.02.003
[18] SASAN M, BABAK S, HASSAN M. A new member of tau-class glutathione S-transferase from barley leaves[J]. Excli Journal, 2009, 8: 190-194.
[19] DROOG F, HOOYKAAS P, VAN DER ZAAL B J. 2, 4-dichlorophenoxyacetic acid and related chlorinated compounds inhibit two auxin-regulated type-III tobacco glutathione S-transferases[J]. Plant Physiology, 1995, 107(4): 1139-1146. doi: 10.1104/pp.107.4.1139
[20] ÖZTETIK E. A tale of plant glutathione S-transferases: Since 1970[J]. The Botanical Review, 2008, 74(3): 419-437. doi: 10.1007/s12229-008-9013-9
[21] LIGHT G G, MAHAN J R, ROXAS V P, et al. Transgenic cotton (Gossypium hirsutum L.) seedlings expressing a tobacco glutathione S-transferase fail to provide improved stress tolerance[J]. Planta, 2005, 222(2): 346-354. doi: 10.1007/s00425-005-1531-7
[22] DIXON D P, CUMMINS I, COLE D J, et al. Glutathione-mediated detoxification systems in plants[J]. Current Opinion in Plant Biology, 1998, 1(3): 258-266. doi: 10.1016/S1369-5266(98)80114-3
[23] NUTRICATI E, MICELI A, BLANDO F, et al. Characterization of two Arabidopsis thaliana glutathione S-transferases[J]. Plant Cell Reports, 2006, 25(9): 997-1005. doi: 10.1007/s00299-006-0146-1
[24] PANTELIDES I S, TJAMOS S E, PAPLOMATAS E J. Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae[J]. Molecular Plant Pathology, 2010, 11(2): 191-202. doi: 10.1111/j.1364-3703.2009.00592.x
[25] BELA K, HORVÁTH E, GALLÉ Á, et al. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses[J]. Journal of Plant Physiology, 2015, 176: 192-201. doi: 10.1016/j.jplph.2014.12.014
[26] BIELACH A, HRTYAN M, TOGNETTI V. Plants under stress: Involvement of auxin and cytokinin[J]. International Journal of Molecular Sciences, 2017, 18(7): 1427. doi: 10.3390/ijms18071427.
[27] LIU J P, LUO X Y, SHAFF J, et al. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance[J]. The Plant Journal, 2012, 71(2): 327-337. doi: 10.1111/j.1365-313X.2012.04994.x
[28] MANGEON A, PARDAL R, MENEZES-SALGUEIRO A D, et al. AtGRP3 is implicated in root size and aluminum response pathways in Arabidopsis[J]. PLoS One, 2016, 11(3): e0150583. doi: 10.1371/journal.pone.0150583.
[29] MINERVA B. Five decades with glutathione and the GSTome[J]. Journal of Biological Chemistry, 2012, 287(9): 6072-6083. doi: 10.1074/jbc.X112.342675
[30] LABROU N E, PAPAGEORGIOU A C, PAVLI O, et al. Plant GSTome: Structure and functional role in xenome network and plant stress response[J]. Current Opinion in Biotechnology, 2015, 32: 186-194. doi: 10.1016/j.copbio.2014.12.024
[31] JHA B, SHARMA A, MISHRA A. Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance[J]. Molecular Biology Reports, 2011, 38(7): 4823-4832. doi: 10.1007/s11033-010-0625-x
[32] TIWARI V, PATEL M K, CHATURVEDI A K, et al. Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress[J]. PLoS One, 2016, 11(2): e0148494. doi: 10.1371/journal.pone.0148494.
[33] XU J, ZHENG A Q, XING X J, et al. Transgenic Arabidopsis plants expressing grape glutathione S-transferase gene (VvGSTF13) show enhanced tolerance to abiotic stress[J]. Biochemistry (Moscow), 2018, 83(6): 755-765. doi: 10.1134/S0006297918060135
[34] SRIVASTAVA D, VERMA G, CHAUHAN A S, et al. Rice (Oryza sativa L.) tau class glutathione S-transferase (OsGSTU30) overexpression in Arabidopsis thaliana modulates a regulatory network leading to heavy metal and drought stress tolerance[J]. Metallomics, 2019, 11(2): 375-389. doi: 10.1039/C8MT00204E
[35] KAMPRANIS S C, DAMIANOVA R, ATALLAH M, et al. A novel plant glutathione S-transferase/peroxidase suppresses bax lethality in yeast[J]. Journal of Biological Chemistry, 2000, 275(38): 29207-29216. doi: 10.1074/jbc.M002359200
[36] LOYALL L, UCHIDA K, BRAUN S, et al. Glutathione and a UV light-induced glutathione S-transferase are involved in signaling to chalcone synthase in cell cultures[J]. The Plant Cell, 2000, 12(10): 1939-1950.
[37] MUELLER L A, GOODMAN C D, SILADY R A, et al. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein[J]. Plant Physiology, 2000, 123(4): 1561-1570. doi: 10.1104/pp.123.4.1561
[38] WEISERBS K F, JACOBSON J S, BEGG M D, et al. A cross-sectional study of polycyclic aromatic hydrocarbon-DNA adducts and polymorphism of glutathione S-transferases among heavy smokers by race/ethnicity[J]. Biomarkers, 2003, 8(2): 142-155. doi: 10.1080/1354750031000086269
[39] GARCERÁ A, BARRETO L, PIEDRAFITA L, et al. Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-cys thiol transferases[J]. The Biochemical Journal, 2006, 398(2): 187-196. doi: 10.1042/BJ20060034
[40] FEDERICI L, MASULLI M, GIANNI S, et al. A conserved hydrogen-bond network stabilizes the structure of Beta class glutathione S-transferases[J]. Biochemical and Biophysical Research Communications, 2009, 382(3): 525-529. doi: 10.1016/j.bbrc.2009.03.052
[41] LALLEMENT P A, RORET T, TSAN P, et al. Insights into ascorbate regeneration in plants: Investigating the redox and structural properties of dehydroascorbate reductases from Populus trichocarpa[J]. Biochemical Journal, 2016, 473(6): 717-731. doi: 10.1042/BJ20151147
[42] GONZALEZ D, FRAICHARD S, GRASSEIN P, et al. Characterization of a Drosophila glutathione transferase involved in isothiocyanate detoxification[J]. Insect Biochemistry and Molecular Biology, 2018, 95: 33-43. doi: 10.1016/j.ibmb.2018.03.004
[43] TAJC S G, TOLBERT B S, BASAVAPPA R, et al. Direct determination of thiol pKa by isothermal titration microcalorimetry[J]. Journal of the American Chemical Society, 2004, 126(34): 10508-10509. doi: 10.1021/ja047929u
[44] ATKINSON H J, BABBITT P C. Glutathione transferases are structural and functional outliers in the thioredoxin fold[J]. Biochemistry, 2009, 48(46): 11108-11116. doi: 10.1021/bi901180v
[45] OHNO S. Patterns in genome evolution[J]. Current Opinion in Genetics & Development, 1993, 3(6): 911-914.
[46] FORCE A, LYNCH M, PICKETT F B, et al. Preservation of duplicate genes by complementary, degenerative mutations[J]. Genetics, 1999, 151(4): 1531-1545. doi: 10.1093/genetics/151.4.1531
[47] TANAKA K M, TAKAHASI K R, TAKANO-SHIMIZU T. Enhanced fixation and preservation of a newly arisen duplicate gene by masking deleterious loss-of-function mutations[J]. Genetics Research, 2009, 91(4): 267-280. doi: 10.1017/S0016672309000196
[48] 马立功, 孟庆林, 张匀华, 等. 向日葵谷胱甘肽-S-转移酶基因的克隆及抗病功能研究[J]. 中国油料作物学报, 2015, 37(5): 635-643. doi: 10.7505/j.issn.1007-9084.2015.05.007 [49] 韩少怀, 李佳佳, 张璟曜, 等. 大豆GmGST12基因的克隆及表达分析[J]. 大豆科学, 2015, 34(5): 782-788. doi: 10.11861/j.issn.1000-9841.2015.05.0782 [50] 李永生, 方永丰, 李玥, 等. 玉米逆境响应基因ZmGST23克隆和表达分析[J]. 农业生物技术学报, 2016, 24(5): 667-677. [51] 孙兰兰, 麻荣慧, 薛飞, 等. 玉米GST31基因的克隆与表达分析[J]. 作物学报, 2023, 49(10): 2717-2726. -
期刊类型引用(3)
1. 董超,董丽英,薛静,杨雅云,汤翠凤,阿新祥,张斐斐,杨勤忠,戴陆园. 云南三地布朗族聚集区地方稻种资源苗瘟抗性评价及抗性基因鉴定. 广东农业科学. 2025(01): 1-13 .  百度学术
百度学术
2. 于雅洁,曹鹏辉,宋云生,陈飞,乔中英,董明辉,朱勇良,谢裕林,袁彩勇. 江苏太湖地区粳稻稻瘟病抗性基因(Pi2、Pita、Pib)的分布及抗病效应. 江苏农业学报. 2024(05): 769-776 .  百度学术
百度学术
3. 范德佳,王汝琴,陈士强,何震天,张容,韩燕,王建华. 扬辐粳系列水稻新品系稻瘟病抗性基因与穗颈瘟抗性分析. 江苏农业科学. 2024(22): 128-132 .  百度学术
百度学术
其他类型引用(0)



 下载:
下载: