Identification of resistant starch related genes in rice by single segment substitution lines
-
摘要:目的
发掘稻米淀粉合成相关基因SBEIIb、SSIIa和ISA1的高抗性淀粉等位基因。
方法利用分子标记筛选携带SBEIIb、SSIIa或ISA1的单片段代换系(Single segment substitution lines,SSSLs),利用改良的AOAC法测定SSSLs材料的抗性淀粉含量(w),通过Sanger测序分析不同SSSLs的SBEIIb、SSIIa和ISA1基因序列,结合基因型和表型连锁分析,鉴定影响抗性淀粉含量的等位基因。
结果SBEIIb编码区的1个SNP(Ex4-96G/A)引起1个氨基酸的替换(196-Arg/His),从而产生2种等位基因SBEIIb-1和SBEIIb-2。其中,SBEIIb-1的Ex4-96G导致第196位氨基酸为Arg,表现为高抗性淀粉含量,为1.72%。SSIIa第8外显子的2个SNPs(Ex8–334G/A和Ex8–865C/T)引起2个氨基酸替换(604-Gly/Ser和781-Leu/Phe),从而产生3种等位基因SSIIa-1、SSIIa-2和SSIIa-3。其中,SSIIa-1的Ex8–334G和Ex8–865C导致第604和781位氨基酸为Gly和Leu,表现为高抗性淀粉含量,为3.37%。ISA1编码区序列的1个Indel(AGG/---)和1个SNP(Ex17–117C/T)导致第70位氨基酸Glu缺失和第717位氨基酸由Thr变为Met,从而产生3种等位基因ISA1-1、ISA1-2和ISA1-3。其中,ISA1-1编码区的AGG插入和Ex17–117C导致第70和717位氨基酸为Glu和Thr,表现为高抗性淀粉含量,为2.09%。
结论SBEIIb、SSIIa和ISA1是影响水稻抗性淀粉形成的重要基因,这3个基因编码区的SNPs和Indels引起了氨基酸的改变,继而影响了抗性淀粉的含量,鉴定到了3个高抗性淀粉含量的等位基因SBEIIb-1、SSIIa-1和ISA1-1。
Abstract:ObjectiveTo discover the alleles for high resistance starch in starch-synthesis-related genes SBEIIb, SSIIa and ISA1.
MethodThe single segment substitution lines (SSSLs) carrying the starch-synthesis-related genes SBEIIb, SSIIa or ISA1 were detected using molecular markers. Then, the resistant starch contents of the SSSLs were measured using an improved AOAC method. Sanger sequencing and sequence alignment were performed to analyze the sequence variations of SBEIIb, SSIIa and ISA1 in different SSSLs. Through linkage analysis of genotypes and phenotypes, the alleles affecting resistance starch content were identified.
ResultFor SBEIIb gene, a single nucleotide polymorphism (SNP) (Ex4-96G/A) in the coding region results in an amino acid substitution (196-Arg/His), generating two alleles SBEIIb-1 and SBEIIb-2. SBEIIb-1 carrying the Ex4-96G causes Arg at 196th residue, which shows high-resistant starch content of 1.72%. For SSIIa gene, two SNPs (Ex8-334G/A and Ex8-865C/T) in the 8th exon cause two amino acid substitutions (604-Gly/Ser and 781-Leu/Phe), generating three alleles SSIIa-1,SSIIa-2 and SSIIa-3. SSIIa-1 carrying the Ex8-334G and Ex8-865C causes Gly and Leu at 604th and 781th residue respectively, which shows high-resistant starch content of 3.37%. One Indel (AGG/---) and one SNP (C/T) in ISA1 coding region sequence generate three alleles ISA1-1, ISA1-2 and ISA1-3. ISA1-1 carrying AGG-insertion and Ex17-117C causes Glu and Thr at 70th and 717th residue respectively, which shows high-resistant starch content of 2.09%.
ConclusionSBEIIb, SSIIa and ISA1 are key genes regulating resistant starch formation in rice. The SNPs and Indels in coding regions of the three genes lead to amino acid variations, which subsequently affects the resistance starch content. The three alleles SBEIIb-1, SSIIa-1 and ISA1-1 for high-resistant starch content are identified.
-
Keywords:
- Rice /
- Single segment substitution line /
- Resistant starch /
- Allele
-
-
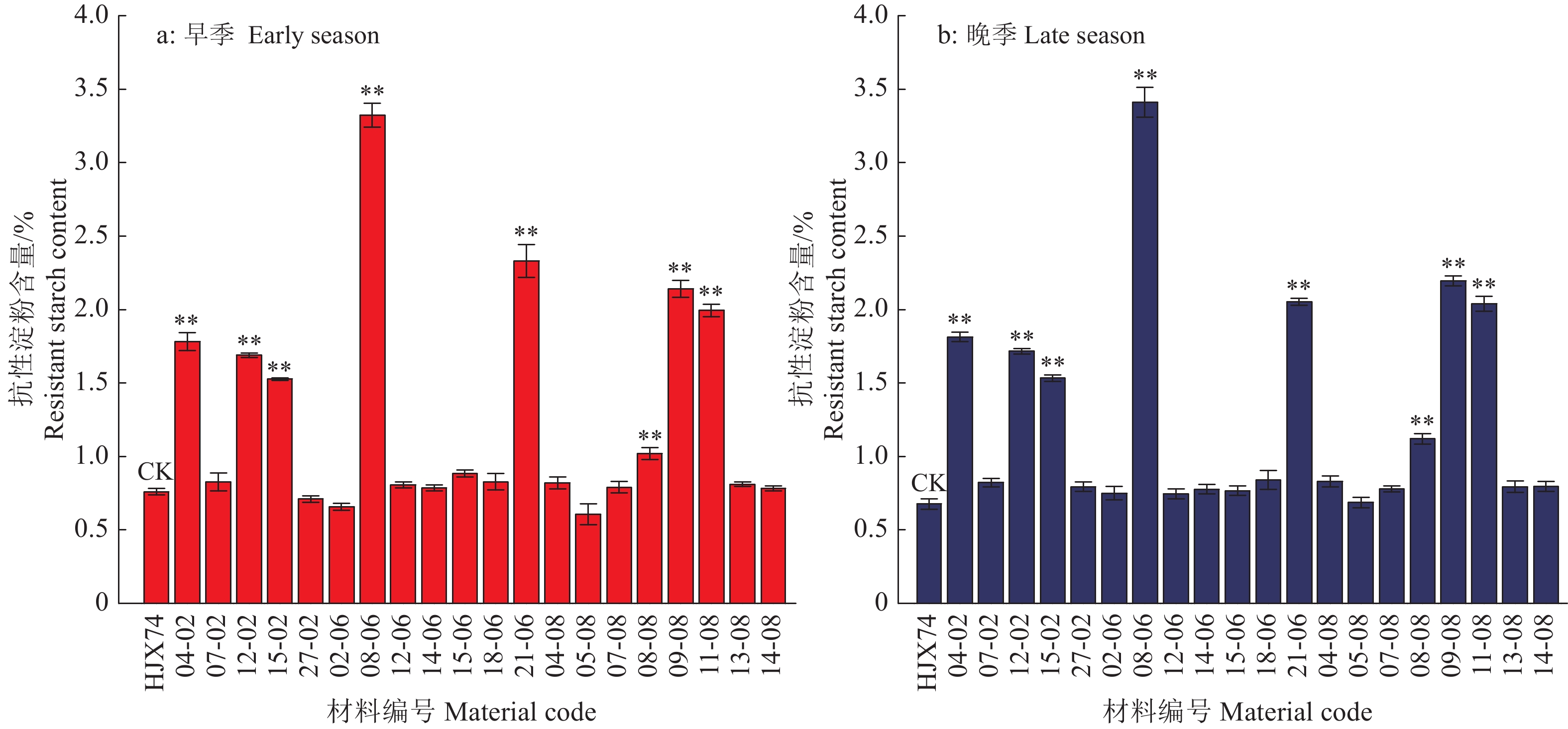
图 2 分别携带SBEIIb、SSIIa和ISA1的SSSLs的抗性淀粉含量
“**”表示SSSLs的抗性淀粉含量与受体亲本HJX74差异显著(P < 0.01,Dunnett’s t 测验,n = 3)
Figure 2. The resistant starch contents of the SSSLs carrying SBEIIb, SSIIa and ISA1, respectively
“**” represents significant difference of the resistant starch between SSSLs and recipient HJX74 (P < 0.01, Dunnett’s t-test, n = 3)
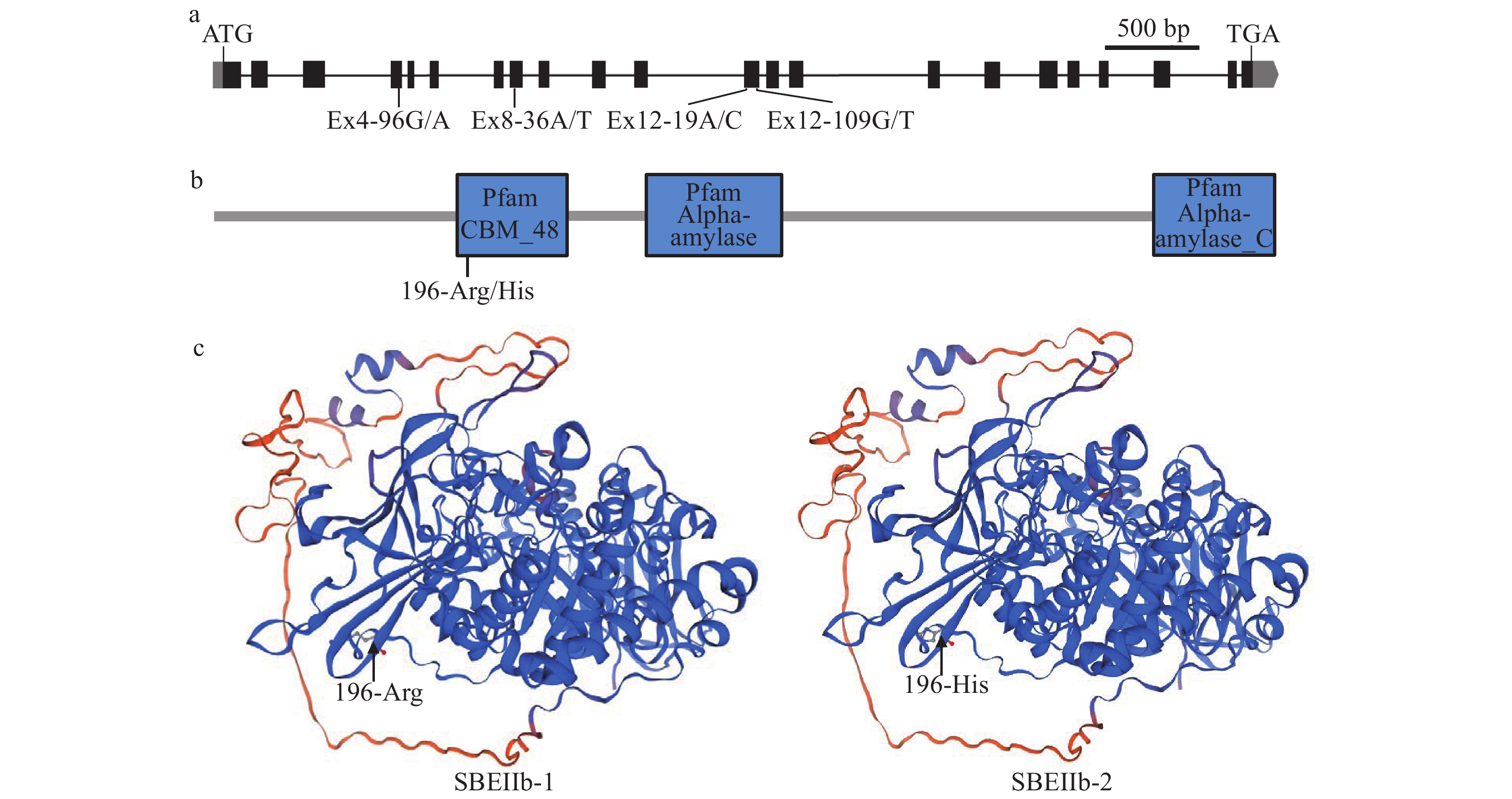
图 3 SSSLs中的SBEIIb等位变异
a:SBEIIb的基因结构示意图;b:SBEIIb蛋白的功能结构域示意图;c:SBEIIb的等位变异蛋白的三维同源性建模
Figure 3. The allelic variations of SBEIIb in SSSLs
a: Gene structure schematic of SBEIIb; b:The schematic illustration of SBEIIb protein functional domains; c: Three-dimensional homology modeling of allelic variant proteins of SBEIIb
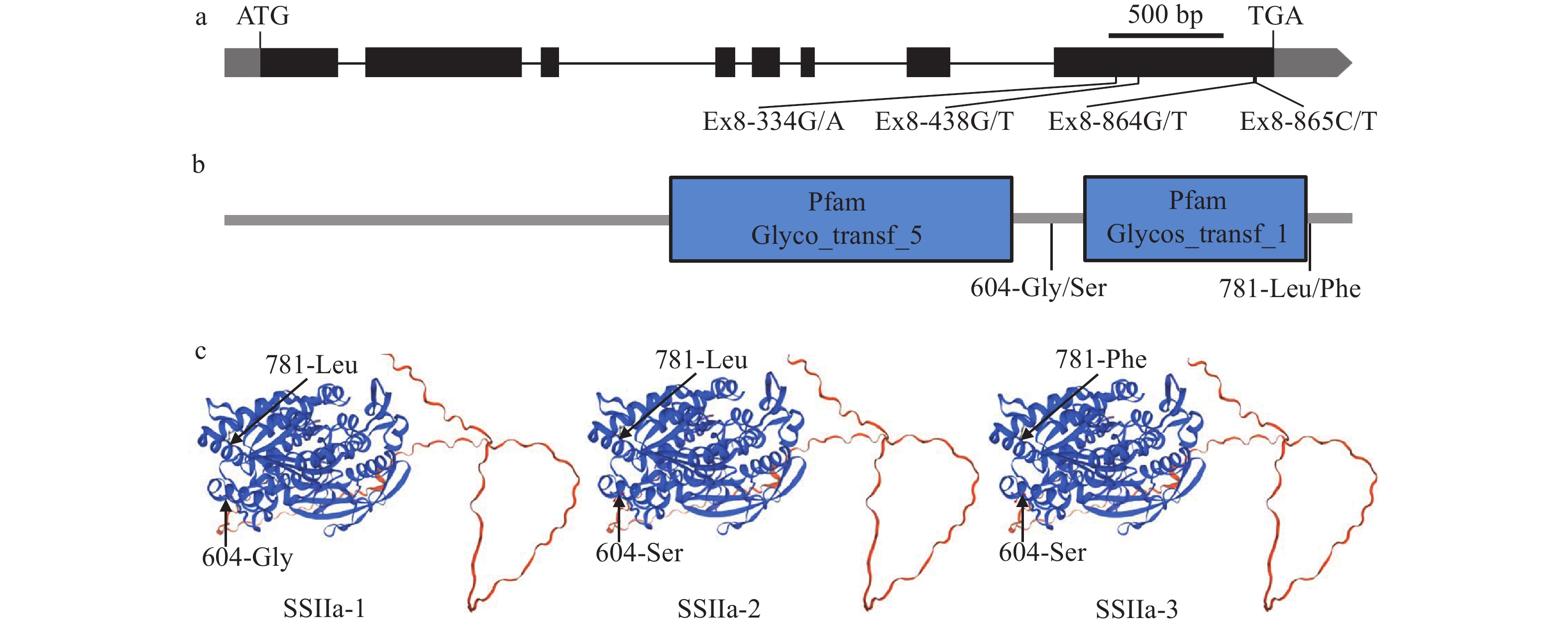
图 4 SSSLs中的SSIIa等位变异
a:SSIIa的基因结构示意图;b:SSIIa蛋白的功能结构域示意图;c:SSIIa的等位变异蛋白的三维同源性建模
Figure 4. The allelic variations of SSIIa in SSSLs
a: Gene structure schematic of SSIIa; b:The schematic illustration of SSIIa protein functional domains; c: Three-dimensional homology modeling of allelic variant proteins of SSIIa
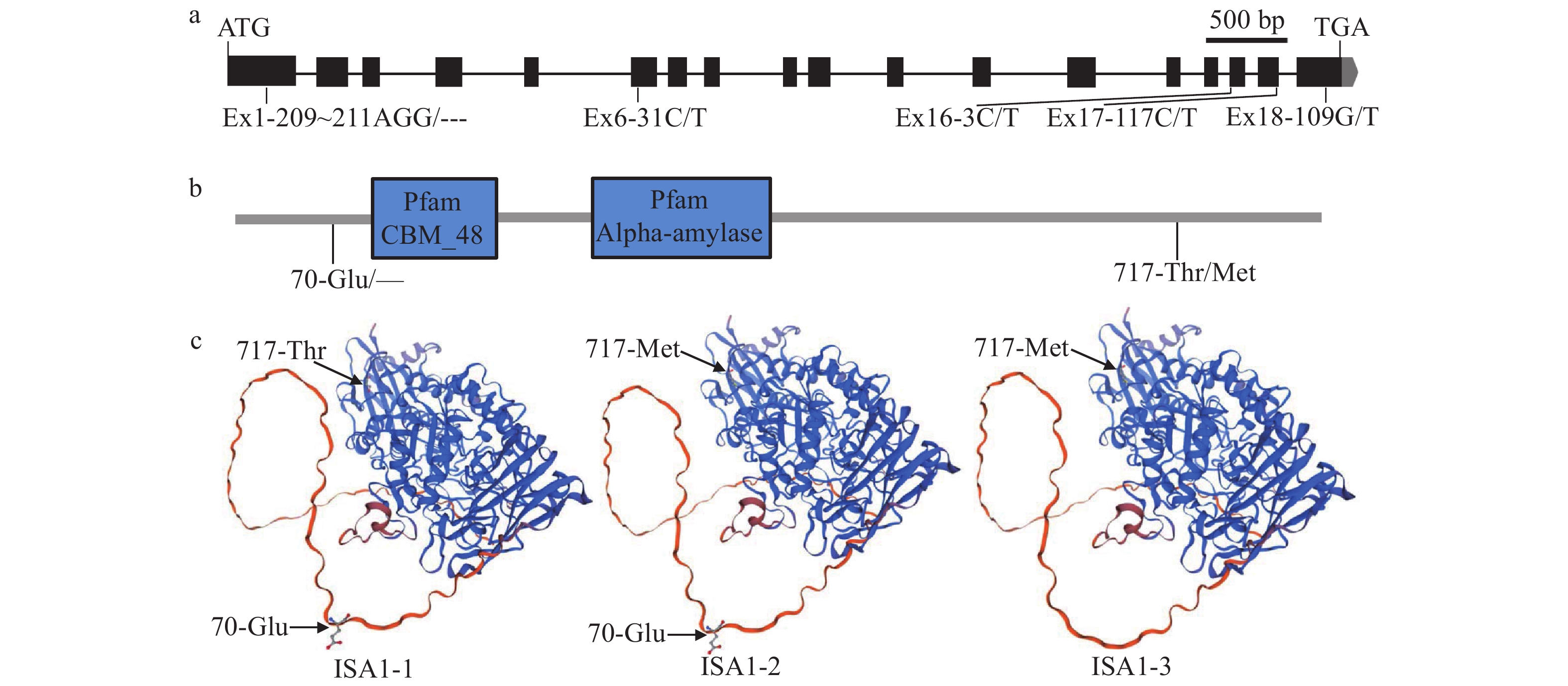
图 5 SSSLs中的ISA1等位变异
a:ISA1的基因结构示意图;b:ISA1蛋白的功能结构域示意图;c:ISA1的等位变异蛋白的三维同源性建模
Figure 5. The allelic variations of ISA1 in SSSLs
a: Gene structure schematic of ISA1; b:The schematic illustration of ISA1 protein functional domains; c: Three-dimensional homology modeling of allelic variant proteins of ISA1
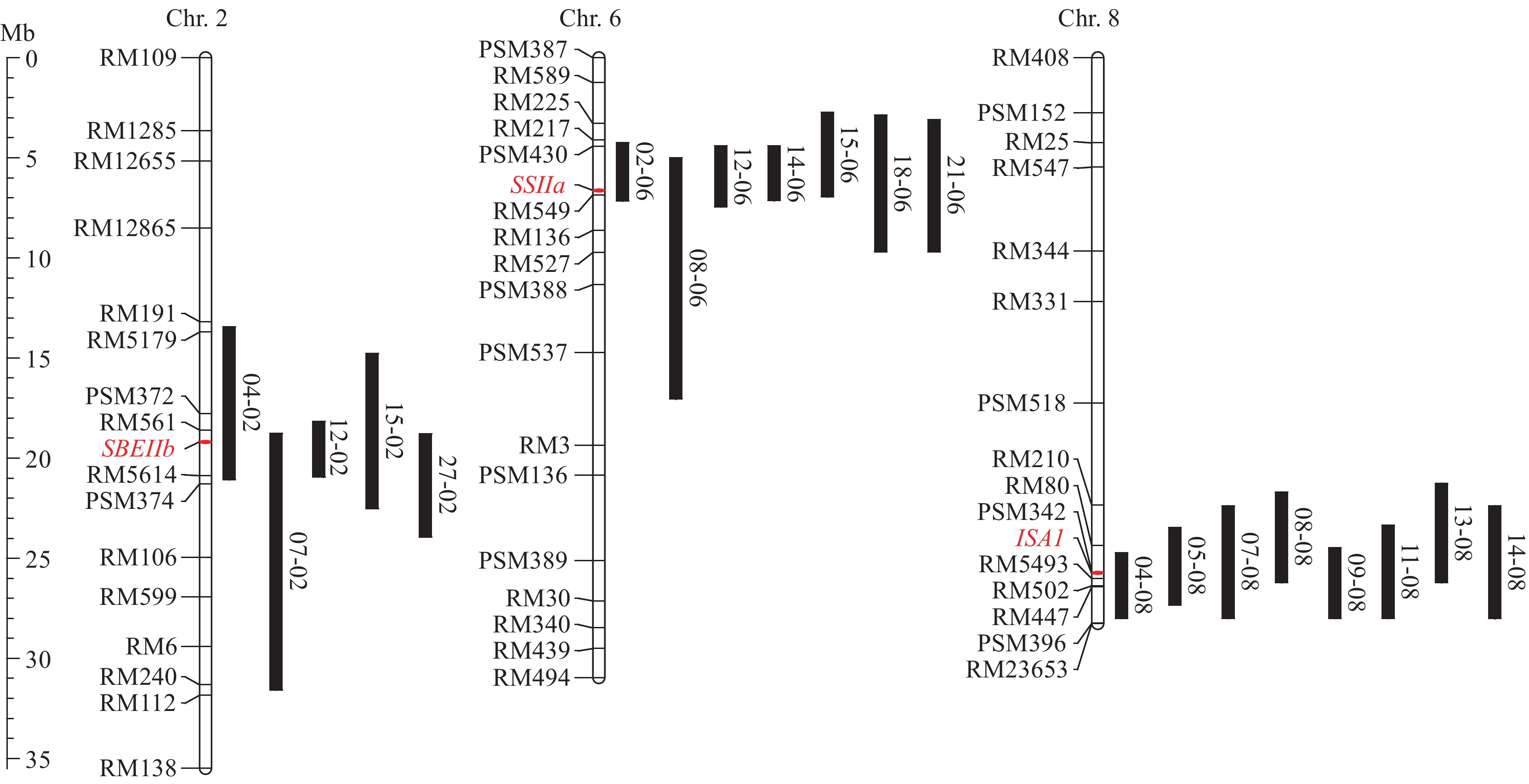
表 1 用于本研究的单片段代换系信息
Table 1 The information of SSSLs used in this study
编号
Code单片段代换系名称
SSSL name染色体
Chr.携带的基因
Carrying gene供体亲本
Donor亚种类型
Subspecies04-02 W04-47-61-02-04 2 SBEIIb BG367 籼稻 Indica 07-02 W07-11-06-04-07-03 2 SBEIIb 苏御糯 Suyunuo 粳稻 Japonica 12-02 W12-09-28-04-01-07-01 2 SBEIIb IR58025B 籼稻 Indica 15-02 W15-12-10-11-07 2 SBEIIb American Jasmine 籼稻 Indica 27-02 W27-18-05-08-01 2 SBEIIb IAPAR9 粳稻 Japonica 02-06 W02-15-01-08-02-05 6 SSIIa Amol 3 籼稻 Indica 08-06 W08-18-09-09-06-02 6 SSIIa IR64 籼稻 Indica 12-06 W12-42-42-08-02-04-02 6 SSIIa IR58025B 籼稻 Indica 14-06 W14-12-03-06-04-08 6 SSIIa 联鉴33 Lianjian33 籼稻 Indica 15-06 W15-06-06-21 6 SSIIa American Jasmine 籼稻 Indica 18-06 W18-11-01-02-06-06-04 6 SSIIa IRAT 261 粳稻 Japonica 21-06 W21-23-36-06-07-04-06 6 SSIIa IR65598 粳稻 Japonica 04-08 W04-10-04-10-07-06 8 ISA1 BG367 籼稻 Indica 05-08 W05-36-75-01-01-06 8 ISA1 籽恢100 Zihui100 籼稻 Indica 07-08 W07-18-05-01-09-03-05 8 ISA1 苏御糯 Suyunuo 粳稻 Japonica 08-08 W08-15-06-05-11 8 ISA1 IR64 籼稻 Indica 09-08 W09-38-60-07-07-11-06 8 ISA1 Basmati 385 籼稻 Indica 11-08 W11-15-08-09-04 8 ISA1 Basmati 370 籼稻 Indica 13-08 W13-30-45-01-10-02-06 8 ISA1 江西丝苗 Jiangxisimiao 籼稻 Indica 14-08 W14-09-06-09-12 8 ISA1 联鉴33 Lianjian33 籼稻 Indica 表 2 SSSLs中的不同SBEIIb等位基因的基因型和表型
Table 2 Genotypes and phenotypes of different SBEIIb alleles in SSSLs
编号
CodeEx4-96
(P.196)Ex8-36
(P.270)Ex12-19
(P.412)Ex12-109
(P.442)单倍型
Haplotype等位基因
Allelew(抗性淀粉)1)/%
Resistant starch content04-02 CGC (Arg) TCA (Ser) ACA (Thr) CTG (Leu) HT-1 SBEIIb-1 1.82±0.03A 12-02 CGC (Arg) TCA (Ser) ACA (Thr) CTG (Leu) HT-1 SBEIIb-1 1.71±0.01A 15-02 CGC (Arg) TCA (Ser) ACA (Thr) CTG (Leu) HT-1 SBEIIb-1 1.63±0.05A HJX74 CAC (His) TCA (Ser) ACA (Thr) CTG (Leu) HT-2 SBEIIb-2 0.72±0.03B 07-02 CAC (His) TCT (Ser) ACC (Thr) CTT (Leu) HT-3 SBEIIb-2 0.82±0.03B 27-02 CAC (His) TCT (Ser) ACC (Thr) CTT (Leu) HT-3 SBEIIb-2 0.75±0.03B 1) 抗性淀粉含量数据为早、晚季的平均值±标准误(n = 6);该列数据后的不同大写字母表示显著差异(P < 0.01,Duncan’s法)
1) The data of resistant starch content are represented as mean ± SE of the early and late seasons (n = 6); Different uppercase letters of this column indicate significant differences (P < 0.01, Duncan’s test)表 3 SSSLs中的不同SSIIa等位基因的基因型和表型
Table 3 Genotypes and phenotypes of different SSIIa alleles in SSSLs
编号
CodeEx8-334
(P.604)Ex8-438
(P.638)Ex8-864
(P.780)Ex8-865
(P.781)单倍型
Haplotype等位基因
Allelew(抗性淀粉)1)/%
Resistant starch content08-06 GGC (Gly) GGG (Gly) GGG (Gly) CTC (Leu) HT-1 SSIIa-1 3.37±0.06A 21-06 AGC (Ser) GGT (Gly) GGG (Gly) CTC (Leu) HT-2 SSIIa-2 2.19±0.08B 02-06 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.71±0.03C 12-06 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.77±0.02C 14-06 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.78±0.02C 15-06 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.82±0.03C 18-06 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.83±0.04C HJX74 AGC (Ser) GGT (Gly) GGT (Gly) TTC (Phe) HT-3 SSIIa-3 0.72±0.03C 1) 抗性淀粉含量数据为早、晚季的平均值±标准误(n = 6);该列数据后的不同大写字母表示显著差异(P < 0.01,Duncan’s法)
1) The data of resistant starch content are represented as mean ± SE of the early and late seasons (n = 6); Different uppercase letters of this column indicate significant differences (P < 0.01, Duncan’s test)表 4 SSSLs中的不同ISA1等位基因的基因型和表型
Table 4 Genotypes and phenotypes of different ISA1 alleles in SSSLs
编号
CodeEx1-209~211
(P.70~71)Ex6-31
(P.336)Ex16-3
(P.353)Ex17-117
(P.717)Ex18-109
(P.777)单倍型
Haplotype等位基因
Allelew(抗性淀粉)1)/%
Resistant starch content09-08 GAGGGT (Glu-Gly) AAC (Asn) GTC (Val) ACG (Thr) CTG (Leu) HT-1 ISA1-1 2.17±0.03A 11-08 GAGGGT (Glu-Gly) AAC (Asn) GTC (Val) ACG (Thr) CTG (Leu) HT-1 ISA1-1 2.02±0.04A 08-08 GAGGGT (Glu-Gly) AAT (Asn) GTT (Val) ATG (Met) CTG (Leu) HT-2 ISA1-2 1.07±0.03B HJX74 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTG (Leu) HT-3 ISA1-3 0.72±0.03C 05-08 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTG (Leu) HT-3 ISA1-3 0.65±0.04C 13-08 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTG (Leu) HT-3 ISA1-3 0.81±0.02C 14-08 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTG (Leu) HT-3 ISA1-3 0.79±0.03C 04-08 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTT (Leu) HT-4 ISA1-3 0.83±0.02C 07-08 G---GT (Gly) AAT (Asn) GTT (Val) ATG (Met) CTT (Leu) HT-4 ISA1-3 0.77±0.02C 1) 抗性淀粉含量数据为早、晚季的平均值±标准误(n = 6);该列数据后的不同大写字母表示显著差异(P < 0.01,Duncan’s法)
1) The data of resistant starch content are represented as mean ± SE of the early and late seasons (n = 6); Different uppercase letters of this column indicate significant differences (P < 0.01, Duncan’s test) -
[1] KHUSH G S. What it will take to feed 5.0 billion rice consumers in 2030[J]. Plant Molecular Biology, 2005, 59(1): 1-6. doi: 10.1007/s11103-005-2159-5
[2] ZOU W, BUTARDO V M, TOUTOUNJI M, et al. Harnessing particle disintegration of cooked rice grains for predicting glycaemic index[J]. Carbohydrate Polymers, 2020, 248: 116789. doi: 10.1016/j.carbpol.2020.116789
[3] RAIGOND P, EZEKIEL R, RAIGOND B. Resistant starch in food: A review[J]. Journal of the Science of Food and Agriculture, 2015, 95(10): 1968-1978. doi: 10.1002/jsfa.6966
[4] 胡时开, 胡培松. 功能稻米研究现状与展望[J]. 中国水稻科学, 2021, 35(4): 311-325. [5] 郑宝东, 王琦, 郑亚凤, 等. 抗性淀粉的生物学功效及在食品加工中的应用[J]. 食品科学技术学报, 2015, 33(5): 1-7. doi: 10.3969/j.issn.2095-6002.2015.05.001 [6] 朱平, 孔祥礼, 包劲松. 抗性淀粉在食品中的应用及功效研究进展[J]. 核农学报, 2015, 29(2): 327-336. doi: 10.11869/j.issn.100-8551.2015.02.0327 [7] BAO J, ZHOU X, XU F, et al. Genome-wide association study of the resistant starch content in rice grains[J]. Starch - Stärke, 2017, 69(7/8): 1600343.
[8] 林静, 张云辉, 张所兵, 等. 水稻地方品种高抗性淀粉含量QTL挖掘与定位[J]. 江苏农业科学, 2021, 49(23): 58-61. [9] 魏霞, 徐延浩, 丁保淼, 等. 抗性淀粉及其遗传改良研究进展[J]. 长江大学学报(自然科学版), 2019, 16(8): 101-107. [10] YANG R, BAI J, FANG J, et al. A single amino acid mutation of OsSBEIIb contributes to resistant starch accumulation in rice[J]. Breeding Science, 2016, 66(4): 481-489. doi: 10.1270/jsbbs.16037
[11] SHEN L, LI J, LI Y. Resistant starch formation in rice: Genetic regulation and beyond[J]. Plant Communications, 2022, 3(3): 100329. doi: 10.1016/j.xplc.2022.100329
[12] SUN Y, JIAO G, LIU Z, et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes[J]. Frontiers in Plant Science, 2017, 8: 298.
[13] ZHANG G, CHENG Z, ZHANG X, et al. Double repression of soluble starch synthase genes SSIIa and SSIIIa in rice (Oryza sativa L. ) uncovers interactive effects on the physicochemical properties of starch[J]. Genome, 2011, 54(6): 448-459. doi: 10.1139/g11-010
[14] ZHOU H, WANG L, LIU G, et al. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(45): 12844-12849.
[15] PAN L, CHEN F, YANG Y, et al. The underlying starch structures of rice grains with different digestibilities but similarly high amylose contents[J]. Food Chemistry, 2022, 379: 132071. doi: 10.1016/j.foodchem.2022.132071
[16] LEHMANN U, ROBIN F. Slowly digestible starch–its structure and health implications: A review[J]. Trends in Food Science & Technology, 2007, 18(7): 346-355.
[17] RAMADOSS B R, GANGOLA M P, AGASIMANI S, et al. Starch granule size and amylopectin chain length influence starch in vitro enzymatic digestibility in selected rice mutants with similar amylose concentration[J]. Journal of Food Science and Technology, 2019, 56(1): 391-400. doi: 10.1007/s13197-018-3500-8
[18] 姚姝, 张亚东, 路凯, 等. 水稻可溶性淀粉合成酶基因SSIIa和SSⅢa的功能、等位变异及其互作研究进展[J]. 中国水稻科学, 2022, 36(3): 227-236. [19] 方结红, 张明洲, 刘军, 等. 水稻ISA1基因的克隆与分析[J]. 安徽农业科学, 2010, 38(9): 4440-4441. [20] CHAO S F, CAI Y C, FENG B B, et al. Editing of rice isoamylase gene ISA1 provides insights into its function in starch formation[J]. Rice Science, 2019, 26(2): 77-87. doi: 10.1016/j.rsci.2018.07.001
[21] FUJITA N, KUBO A, SUH D, et al. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm[J]. Plant and Cell Physiology, 2003, 44(6): 607-618. doi: 10.1093/pcp/pcg079
[22] 张桂权. 基于SSSL文库的水稻设计育种平台[J]. 遗传, 2019, 41(8): 754-760. [23] KAWAGOE Y, KUBO A, SATOH H, et al. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm[J]. The Plant Journal, 2005, 42(2): 164-174. doi: 10.1111/j.1365-313X.2005.02367.x
[24] SAWADA T, ITOH M, NAKAMURA Y. Contributions of three starch branching enzyme isozymes to the fine structure of amylopectin in rice endosperm[J]. Frontiers in Plant Science, 2018, 9: 1536. doi: 10.3389/fpls.2018.01536
[25] MIURA S, KOYAMA N, CROFTS N, et al. Generation and starch characterization of non-transgenic BEI and BEIIb double mutant rice (Oryza sativa) with ultra-high level of resistant starch[J]. Rice, 2021, 14(1): 3. doi: 10.1186/s12284-020-00441-0
[26] GUO D, LING X, ZHOU X, et al. Evaluation of the quality of a high-resistant starch and low-glutelin rice (Oryza sativa L.) Generated through CRISPR/Cas9-mediated targeted mutagenesis[J]. Journal of Agricultural and Food Chemistry, 2020, 68(36): 9733-9742. doi: 10.1021/acs.jafc.0c02995
[27] YANG R, SUN C, BAI J, et al. A putative gene sbe3-rs for resistant starch mutated from SBE3 for starch branching enzyme in rice (Oryza sativa L. )[J]. PLoS One, 2012, 7(8): e43026. doi: 10.1371/journal.pone.0043026
[28] BUTARDO V M, FITZGERALD M A, BIRD A R, et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing[J]. Journal of Experimental Botany, 2011, 62(14): 4927-4941. doi: 10.1093/jxb/err188
[29] GAO Z, ZENG D, CHENG F, et al. ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice[J]. Journal of Integrative Plant Biology, 2011, 53(9): 756-765.
[30] CHEN Z, LU Y, FENG L, et al. Genetic dissection and functional differentiation of ALKa and ALKb, two natural alleles of the ALK/SSIIa gene, responding to low gelatinization temperature in rice[J]. Rice, 2020, 13(1): 39. doi: 10.1186/s12284-020-00393-5
[31] YOU H, LIANG C, ZHANG O, et al. Variation of resistant starch content in different processing types and their starch granules properties in rice[J]. Carbohydrate Polymers, 2022, 276: 118742. doi: 10.1016/j.carbpol.2021.118742
[32] NAKAMURA Y, FRANCISCO P B, HOSAKA Y, et al. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties[J]. Plant Molecular Biology, 2005, 58(2): 213-227. doi: 10.1007/s11103-005-6507-2
[33] 张风琴, 于雪然, 李玲, 等. 基于高密度遗传图谱对水稻抗性淀粉QTL定位及分析[J]. 植物遗传资源学报, 2023, 24(4): 1075-1084.



 下载:
下载: