Functional analysis of OsRAC3 regulating seed traits in rice
-
摘要:目的
RAC/ROPs是一类植物特有的小G蛋白,作为分子开关参与众多的植物信号转导途径。OsRAC3通过与细胞分裂素信号组分互作,调控水稻Oryza sativa L.冠根发育,但是否影响水稻地上部分的性状尚不清楚。本研究主要分析OsRAC3对水稻穗发育以及产量性状的影响。
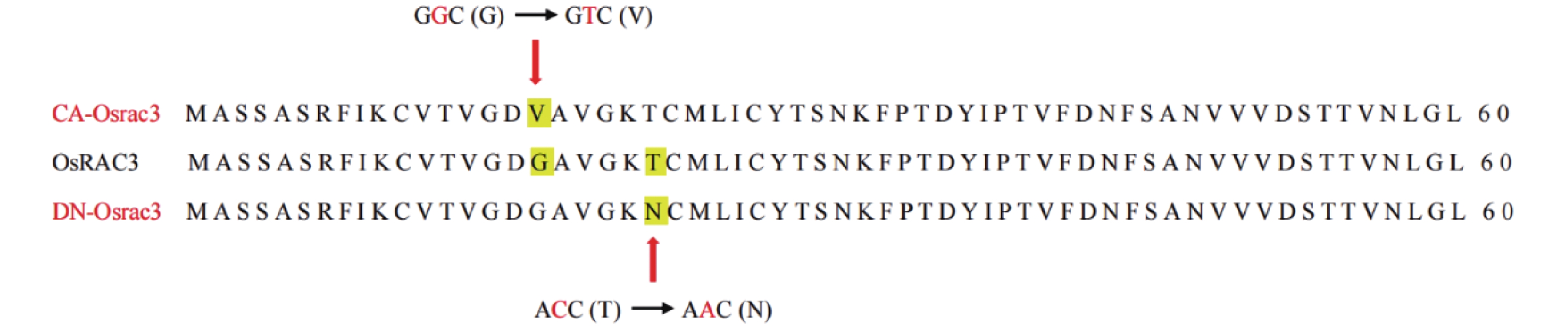
方法对水稻OsRAC3promoter::GUS转基因植株的花序组织进行GUS染色,分析OsRAC3的表达模式。分析转基因材料持续激活型突变体(CA-osrac3)和显性失活型突变体(DN-osrac3)2种试验材料的每穗粒数、粒长、粒宽、千粒质量等主要农艺性状。
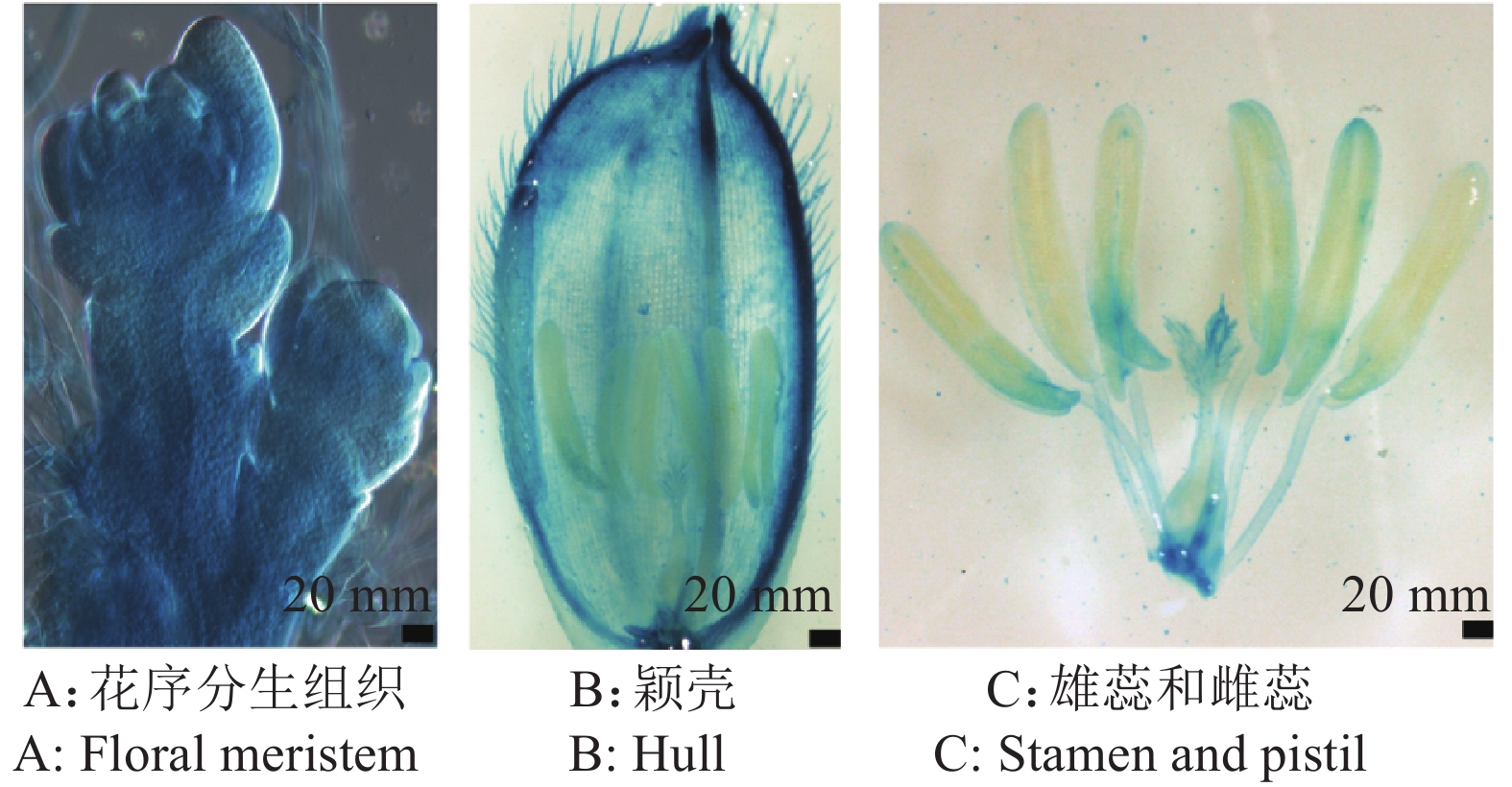
结果OsRAC3在花序分生组织、雄蕊和雌蕊中强烈表达;CA-osrac3株系的穗粒数减少,籽粒更加饱满,粒长、粒宽、千粒质量均显著高于野生型;DN-osrac3株系的穗分枝数少且育性差,籽粒粒长、粒宽、千粒质量则显著低于野生型。
结论OsRAC3影响水稻穗的发育过程,持续激活型突变体CA-osrac3促进水稻籽粒的发育;OsRAC3在水稻高产育种和遗传改良中具有重要的应用价值。
Abstract:ObjectiveRAC/ROPs are a class of plant-specific small G proteins, which are involved in multiple signaling pathways as molecular switches. OsRAC3 regulates rice (Oryza sativa L.) crown root development by interacting with cytokinin signaling components. However whether it affects traits in the aboveground parts of rice remains unclear. This study mainly aims to analyze the effects of OsRAC3 on spike development and yield traits of rice.
MethodThe inflorescence tissue of rice OsRAC3promoter::GUS transgenic plants were stained with GUS to analyze the expression pattern of OsRAC3. We also analyzed the main agronomic traits such as number of grains per spike, grain length, grain width and thousand grain weight of two experimental materials, constitutively activated mutant (CA-osrac3) and dominant negative mutant (DN-osrac3).
ResultOsRAC3 was strongly expressed in floral meristems, stamen and pistill. CA-osrac3 had fewer grains but its grain length, grain width and thousand grain weight were significantly higher than those of wild-type. DN-osrac3 had a low number of spike branches and poor fertility, while the grain length, grain width and thousand grain weight were significantly lower than those of wild-type.
ConclusionOsRAC3 affects the developmental process of rice spikes, and the constitutively activated mutant CA-osrac3 promotes the growth of rice seeds. OsRAC3 has important applications in high-yield breeding and genetic improvement of rice.
-
Keywords:
- Oryza sativa L. /
- RAC/ROP /
- OsRAC3 /
- Spike /
- Grain
-
-
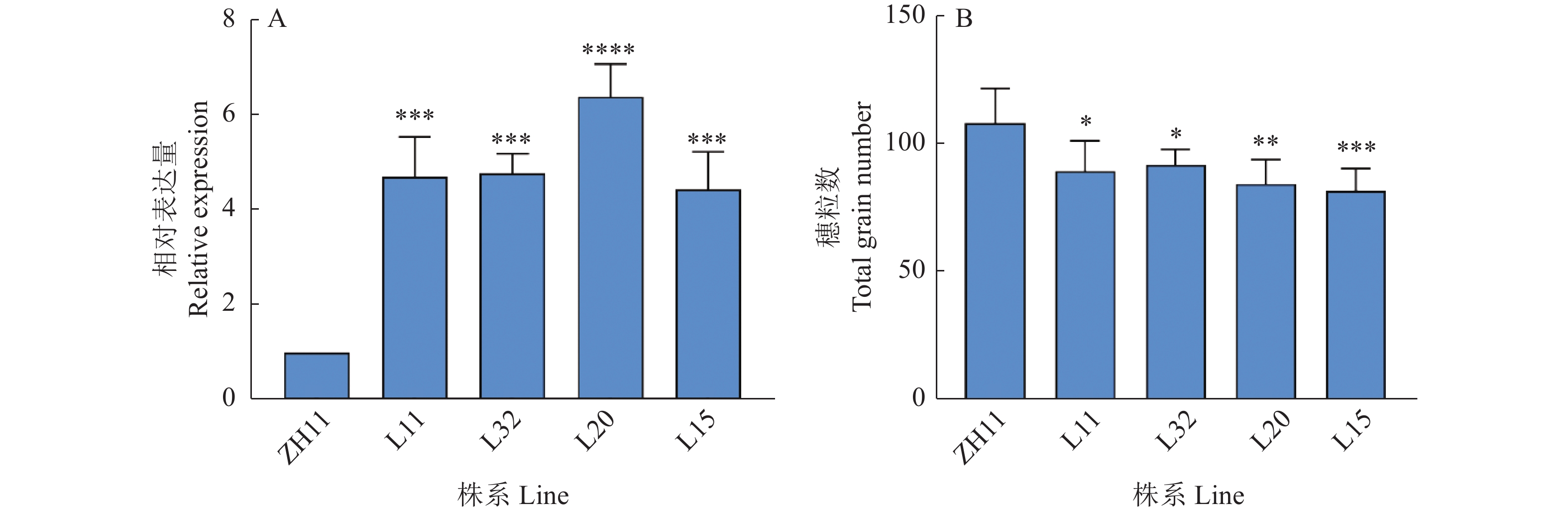
图 3 CA-osrac3、DN-osrac3在野生型ZH11以及突变体CA-osrac3、DN-osrac3中的表达情况
L11、L32为CA-osrac3株系,L20、L15为DN-osrac3株系;“*”“**”“***”“****”分别表示突变体株系与野生型在P<0.05、P<0.01、P<0.001、P<0.000 1水平差异显著(t检验)
Figure 3. Expression of CA-osrac3, DN-osrac3 in wild-type ZH11 and mutant lines CA-osrac3, DN-osrac3
L11 and L32 are CA-osrac3 lines, L20 and L15 are DN-osrac3 lines; “*” “**” “***” “****” indicate that the mutant strains differed from the wild-type respectively at P<0.05, P<0.01, P<0.001, P<0.0001 levels of significant difference (t test)
图 5 野生型ZH11以及突变体CA-osrac3、DN-osrac3籽粒特征分析
L11、L32为CA-osrac3株系,L20、L15为DN-osrac3株系;“***”“****”分别表示突变体株系与野生型在P<0.001、P<0.000 1水平差异显著(t检验)
Figure 5. Grain analysis of wild-type ZH11 and mutant lines CA-osrac3, DN-osrac3
L11 and L32 are CA-osrac3 lines, L20 and L15 are DN-osrac3 lines; “***” “****” indicate that the mutant lines differed from the wild-type respectively at P<0.001, P<0.0001 levels of significant difference (t test)
-
[1] ROSEGRANT M W, CLINE S A. Global food security: Challenges and policies[J]. Science, 2003, 302(5652): 1917-1919. doi: 10.1126/science.1092958
[2] GODFRAY H C J, BEDDINGTON J R, CRUTE I R, et al. Food security: The challenge of feeding 9 billion people[J]. Science, 2010, 327(5967): 812-818. doi: 10.1126/science.1185383
[3] ZHOU Y, TAO Y, YUAN Y, et al. Characterisation of a novel quantitative trait locus, GN4-1, for grain number and yield in rice (Oryza sativa L. )[J]. Theoretical and Applied Genetics, 2018, 131(3): 637-648. doi: 10.1007/s00122-017-3025-y
[4] SAKAMOTO T, MATSUOKA M. Identifying and exploiting grain yield genes in rice[J]. Current Opinion in Plant Biology, 2008, 11(2): 209-214. doi: 10.1016/j.pbi.2008.01.009
[5] LI S, ZHAO B, YUAN D, et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(8): 3167-3172. doi: 10.1073/pnas.1300359110
[6] 吴琳华, 陈文丰. 水稻花序结构调控机制研究进展[J]. 广东农业科学, 2022, 49(9): 42-52. doi: 10.16768/j.issn.1004-874X.2022.09.005 [7] 董皓, 李懿星, 宋书锋, 等. 水稻穗粒数相关基因研究进展[J]. 分子植物育种, 2021, 19(18): 6045-6050. doi: 10.13271/j.mpb.019.006045 [8] KOMATSU K, MAEKAWA M, UJIIE S, et al. LAX and SPA: Major regulators of shoot branching in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(20): 11765-11770. doi: 10.1073/pnas.1932414100
[9] IKEDA K, ITO M, NAGASAWAO N, et al. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate[J]. Plant Journal, 2007, 51(6): 1030-1040. doi: 10.1111/j.1365-313X.2007.03200.x
[10] KURAKAWA T, UEDA N, MAEKAWA M, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme[J]. Nature, 2007, 445(7128): 652-655. doi: 10.1038/nature05504
[11] ASHIKARI M, SAKAKIBARA H, LIN S Y, et al. Cytokinin oxidase regulates rice grain production[J]. Science, 2005, 309(5735): 741-745. doi: 10.1126/science.1113373
[12] TAN L, LI X, LIU F, et al. Control of a key transition from prostrate to erect growth in rice domestication[J]. Nature Genetics, 2008, 40(11): 1360-1364. doi: 10.1038/ng.197
[13] JIN J, HUANG W, GAO J, et al. Genetic control of rice plant architecture under domestication[J]. Nature Genetics, 2008, 40(11): 1365-1369. doi: 10.1038/ng.247
[14] HUANG X, QIAN Q, LIU Z, et al. Natural variation at the DEP1 locus enhances grain yield in rice[J]. Nature Genetics, 2009, 41(4): 494-497. doi: 10.1038/ng.352
[15] IKEDA-KAWAKATSU K, YASUNO N, OIKAWA T, et al. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem[J]. Plant Physiology, 2009, 150(2): 736-747. doi: 10.1104/pp.109.136739
[16] JIAO Y, WANG Y, XUE D, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice[J]. Nature Genetics, 2010, 42(6): 536-541. doi: 10.1038/ng.574
[17] OOKAWA T, HOBO T, YANO M, et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield[J]. Nature Communications, 2010, 1: 132. doi: 10.1038/ncomms1132.
[18] MIURA K, IKEDA M, MATSUBARA A, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice[J]. Nature Genetics, 2010, 42(6): 545-549.
[19] WU H, HAZAK O, CHEUNG A Y, et al. RAC/ROP GTPases and auxin signaling[J]. Plant Cell, 2011, 23(4): 1208-1218. doi: 10.1105/tpc.111.083907
[20] FEIGUELMAN G, FU Y, YALOVSKY S. ROP GTPases structure-function and signaling pathways[J]. Plant Physiology, 2018, 176(1): 57-79. doi: 10.1104/pp.17.01415
[21] 刘礼瑶, 赵博, 熊仁次. 植物小G蛋白ROPs生物学功能研究进展[J]. 现代园艺, 2023, 46(9): 37-40. doi: 10.3969/j.issn.1006-4958.2023.09.014 [22] 吕畅, 周利明. ROP信号途径在细胞极性发育中的研究进展[J]. 农业与技术, 2023, 43(6): 10-13. doi: 10.19754/j.nyyjs.20230330003 [23] 郭亚如, 陈欣, 黄俊骏. ROP蛋白在植物生长发育及逆境响应中的作用研究进展[J]. 河南农业科学, 2021, 50(11): 1-5. doi: 10.15933/j.cnki.1004-3268.2021.11.001 [24] 廖恒毅, 王若霖, 黄进. ROPs: 植物细胞内多种信号通路的分子开关[J]. 中国生物化学与分子生物学报, 2022, 38(3): 271-283. [25] ETIENNE-MANNEVILLE S, HALL A. Rho GTPases in cell biology[J]. Nature, 2002, 420(6916): 629-635. doi: 10.1038/nature01148
[26] BERKEN A, WITTINGHOFER A. Structure and function of Rho-type molecular switches in plants[J]. Plant Physiology and Biochemistry, 2008, 46(3): 380-393. doi: 10.1016/j.plaphy.2007.12.008
[27] KOST B, LEMICHEZ E, SPIELHOFER P, et al. Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth[J]. Journal of Cell Biology, 1999, 145(2): 317-330. doi: 10.1083/jcb.145.2.317
[28] FU Y, WU G, YANG Z. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes[J]. Journal of Cell Biology, 2001, 152(5): 1019-1032. doi: 10.1083/jcb.152.5.1019
[29] WU G, GU Y, LI S, et al. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets[J]. Plant Cell, 2001, 13(12): 2841-2856.
[30] MOLENDIJK A J, BISCHOFF F, RAJENDRAKUMAR C S, et al. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth[J]. The EMBO Journal, 2001, 20(11): 2779-2788. doi: 10.1093/emboj/20.11.2779
[31] JONES M A, SHEN J J, FU Y, et al. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth[J]. Plant Cell, 2002, 14(4): 763-776. doi: 10.1105/tpc.010359
[32] CRADDOCK C, LAVAGI I, YANG Z. New insights into Rho signaling from plant ROP/Rac GTPases[J]. Trends in Cell Biology, 2012, 22(9): 492-501. doi: 10.1016/j.tcb.2012.05.002
[33] CHOI Y, LEE Y, HWANG J. Arabidopsis ROP9 and ROP10 GTPases differentially regulate auxin and ABA responses[J]. Journal of Plant Biology, 2014, 57(4): 245-254. doi: 10.1007/s12374-014-0029-x
[34] LI H, LIN Y, HEATH R M, et al. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx[J]. Plant Cell, 1999, 11(9): 1731-1742.
[35] CHEN L, SHIOTANI K, TOGASHI T, et al. Analysis of the Rac/Rop small GTPase family in rice: Expression, subcellular localization and role in disease resistance[J]. Plant and Cell Physiology, 2010, 51(4): 585-595. doi: 10.1093/pcp/pcq024
[36] ONO E, WONG H L, KAWASAKI T, et al. Essential role of the small GTPase Rac in disease resistance of rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(2): 759-764. doi: 10.1073/pnas.98.2.759
[37] KIM S H, OIKAWA T, KYOZUKA J, et al. The bHLH Rac immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity[J]. Plant and Cell Physiology, 2012, 53(4): 740-754. doi: 10.1093/pcp/pcs033
[38] ZHANG Y, XIONG Y, LIU R, et al. The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(32): 16121-16126. doi: 10.1073/pnas.1902321116
[39] LIU H, HUANG J, ZHANG X, et al. The RAC/ROP GTPase activator OsRopGEF10 functions in crown root development by regulating cytokinin signaling in rice[J]. Plant Cell, 2022, 35(1): 453-468.
[40] YANG Z B. Small GTPases: Versatile signaling switches in plants[J]. Plant Cell, 2002, 14(S): S375-S388.
[41] SAKAMOTO T, MATSUOKA M. Generating high-yielding varieties by genetic manipulation of plant architecture[J]. Current Opinion in Biotechnology, 2004, 15(2): 144-147. doi: 10.1016/j.copbio.2004.02.003
[42] YIN C, ZHU Y, LI X, et al. Molecular and genetic aspects of grain number determination in rice (Oryza sativa L.)[J]. International Journal of Molecular Sciences, 2021, 22(2): 728. doi: 10.3390/ijms22020728.
[43] LI N, XU R, DUAN P, et al. Control of grain size in rice[J]. Plant Reproduction, 2018, 31(3): 237-251. doi: 10.1007/s00497-018-0333-6
[44] BURR C A, SUN J, YAMBURENKO M V, et al. The HK5 and HK6 cytokinin receptors mediate diverse developmental pathways in rice[J]. Development, 2020, 147(20): dev191734. doi: 10.1242/dev.191734.
[45] LI N, LI Y. Signaling pathways of seed size control in plants[J]. Current Opinion in Plant Biology, 2016, 33: 23-32. doi: 10.1016/j.pbi.2016.05.008



 下载:
下载: