Study of Arabidopsis H3K27 methyltransferase CLF responding to ambient temperature and involving in temperature morphogenesis
-
摘要:目的
探索拟南芥H3K27甲基转移酶CURLY LEAF(CLF)在温度形态建成中的作用。
方法在不同温度条件(22和16 ℃)下,对拟南芥Arabidopsis野生型Col-0和突变体clf-29进行表型分析和转录组分析,筛选差异表达基因。
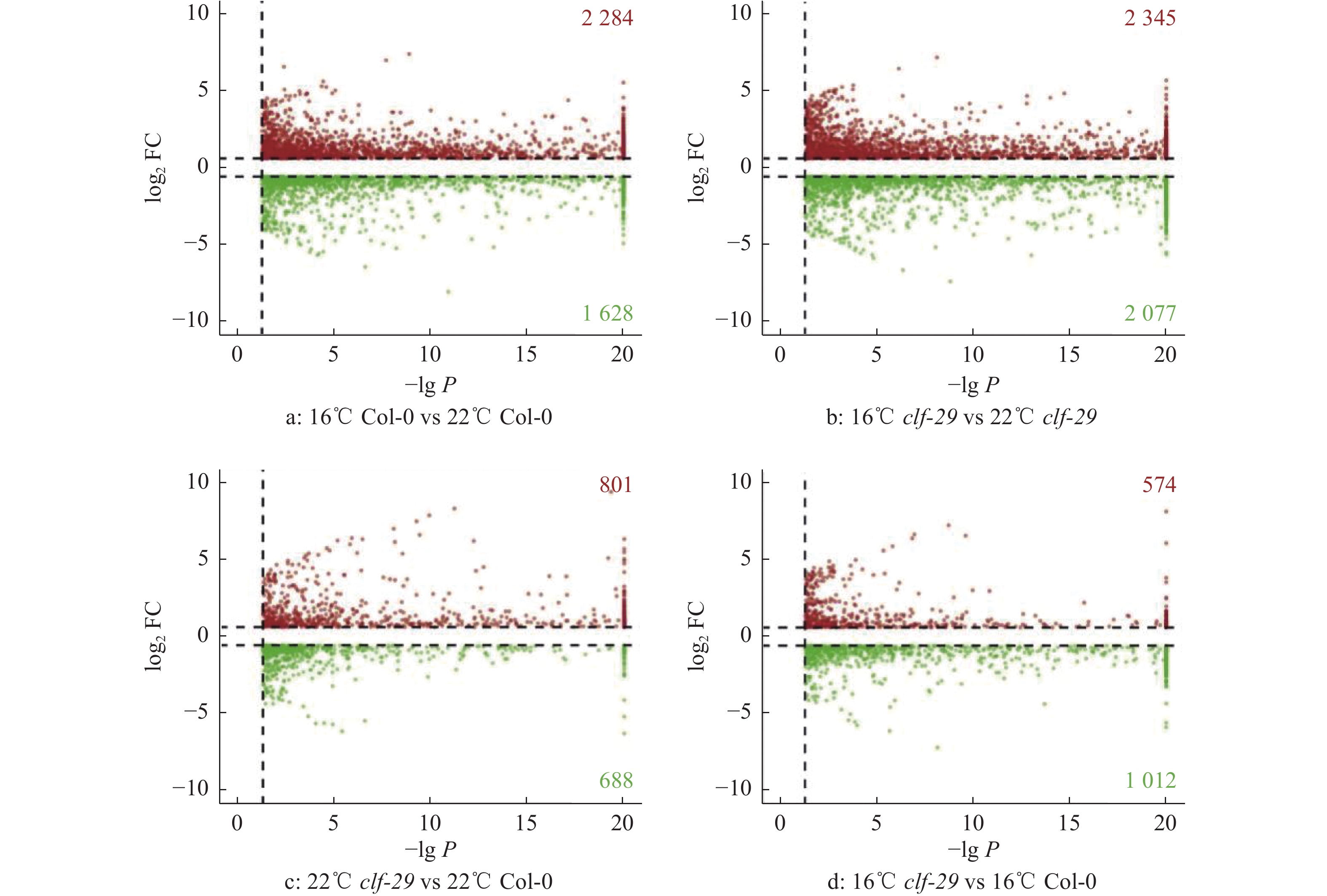
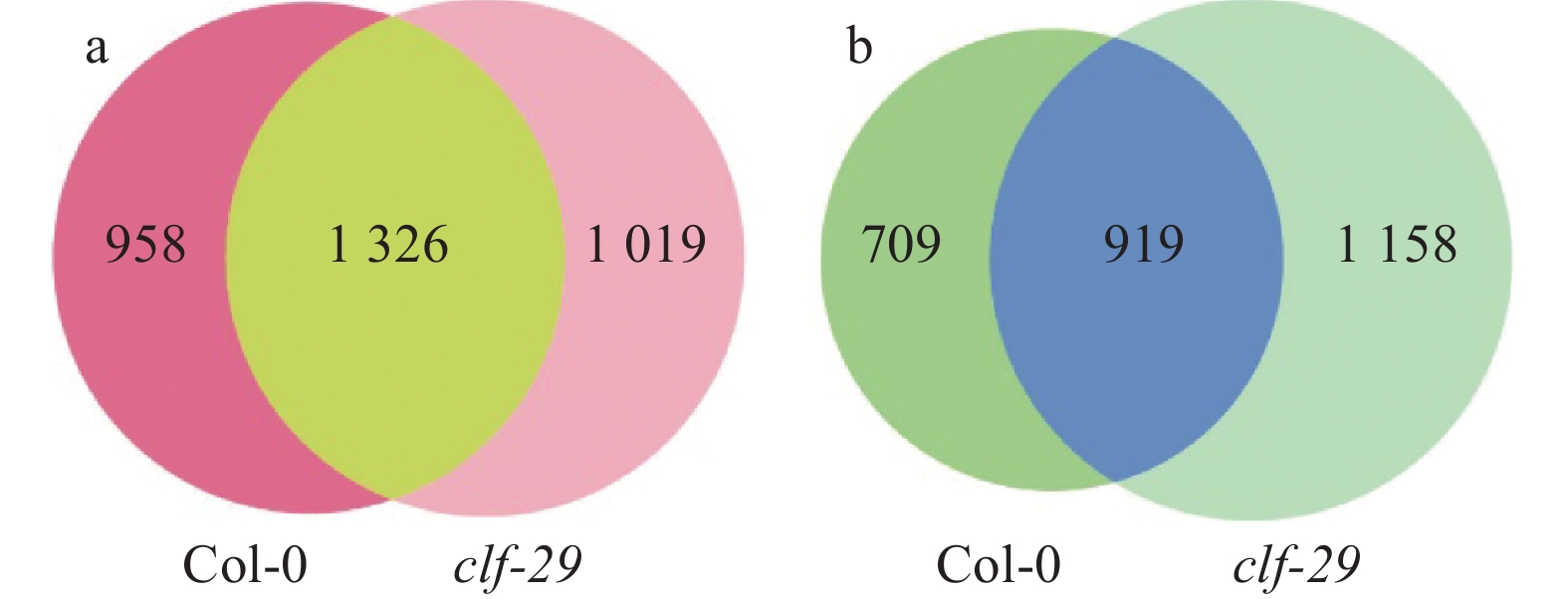
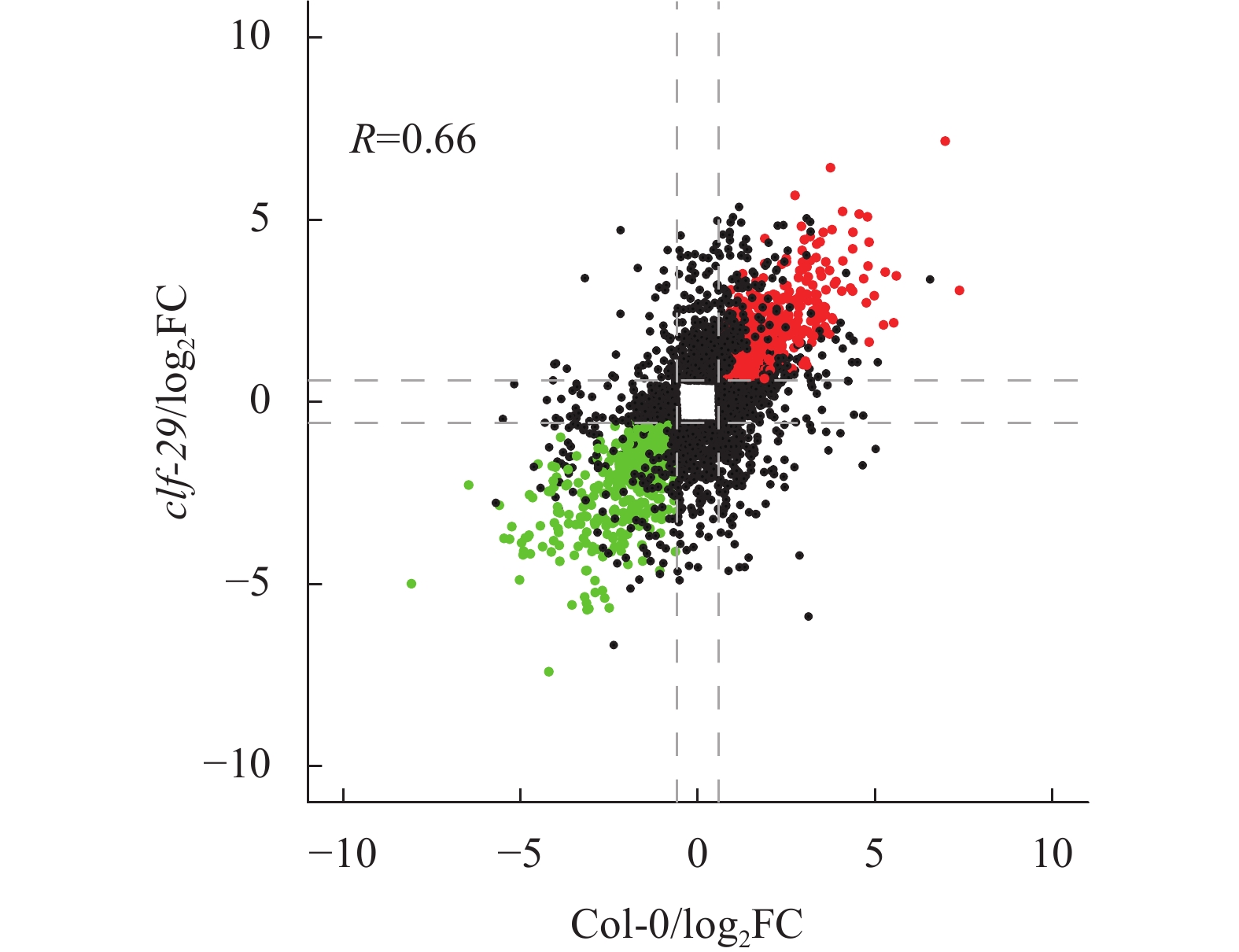
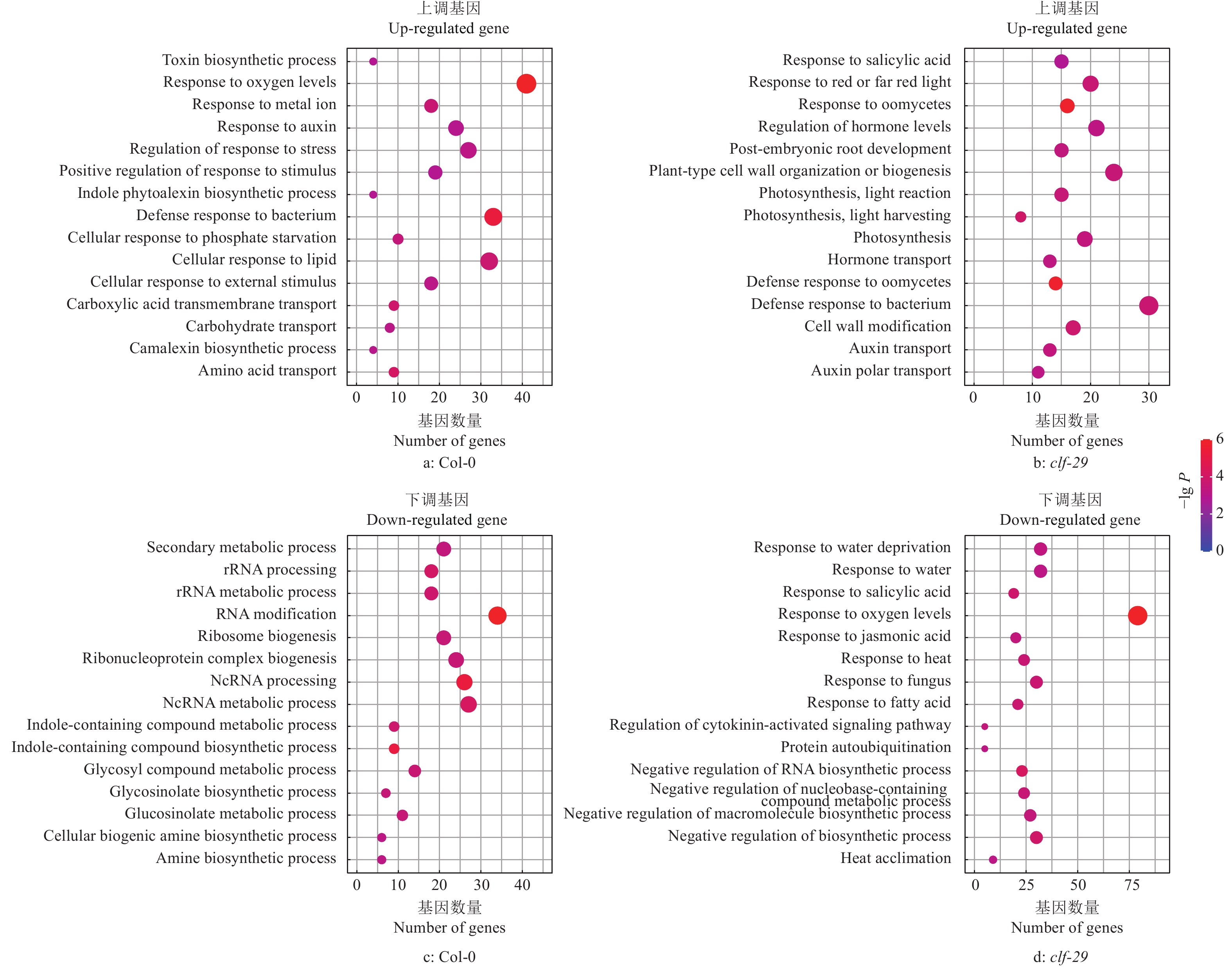
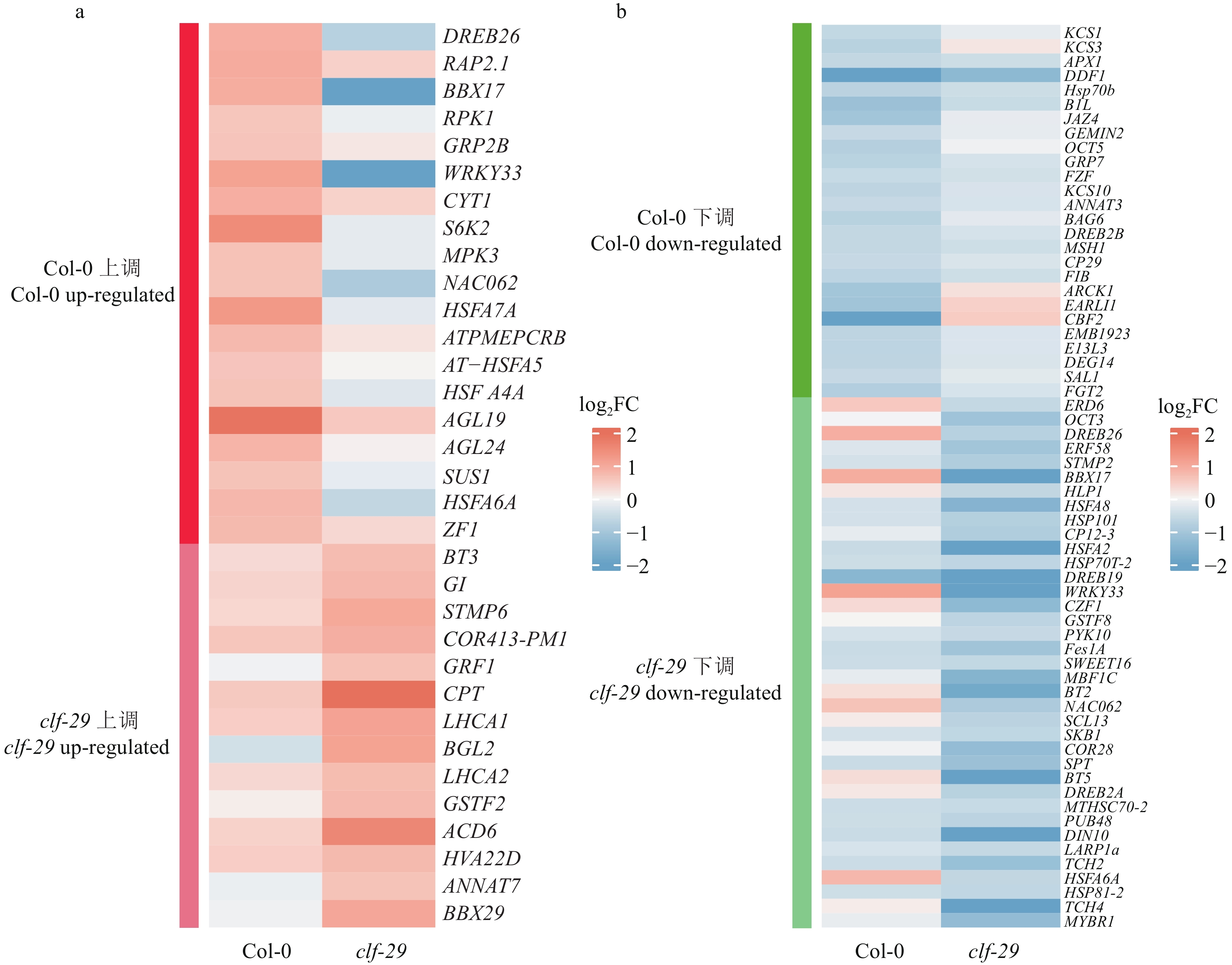
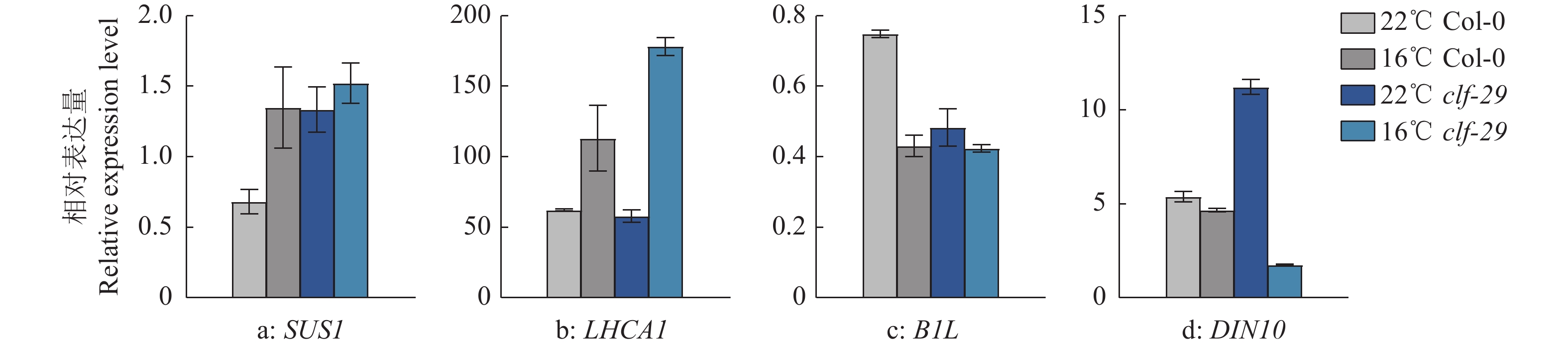
结果在不同温度条件下,clf-29表现出显著的表型差异,相较于22 ℃,16 ℃时clf-29和Col-0的表型差异更小。转录组分析发现CLF的缺失会导致大量基因表达差异,并将其分为4种类型(仅在Col-0显著上调、下调,仅在clf-29突变体显著上调、下调),包含96个温度响应基因。
结论拟南芥表观遗传调控因子CLF响应环境温度,并参与温度形态建成。
Abstract:ObjectiveTo explore the role of Arabidopsis H3K27 methyltransferase CURLY LEAF (CLF) in temperature morphogenesis.
MethodThe differentially expressed genes were screened by phenotypic analysis and transcriptome analysis of Arabidopsis wild type Col-0 and mutant clf-29 under different temperatures of 22 and 16 ℃.
Resultclf-29 showed significant phenotypic differences under different temperatures, there was less difference between clf-29 and Col-0 at 16 ℃ than at 22 ℃. Transcriptome analysis found that deletion of CLF led to expression changes of a large number of genes, which were divided into four types (significantly up-regulated/down-regulated only in Col-0, significantly up-regulated/down-regulated only in clf-29 mutant), containing 96 temperature responsive genes.
ConclusionArabidopsis epigenetic regulator CLF responds to ambient temperature and is involved in temperature morphogenesis.
-
Keywords:
- Arabidopsis /
- Methyltransferase /
- H3K27me3 /
- Temperature morphogenesis
-
-
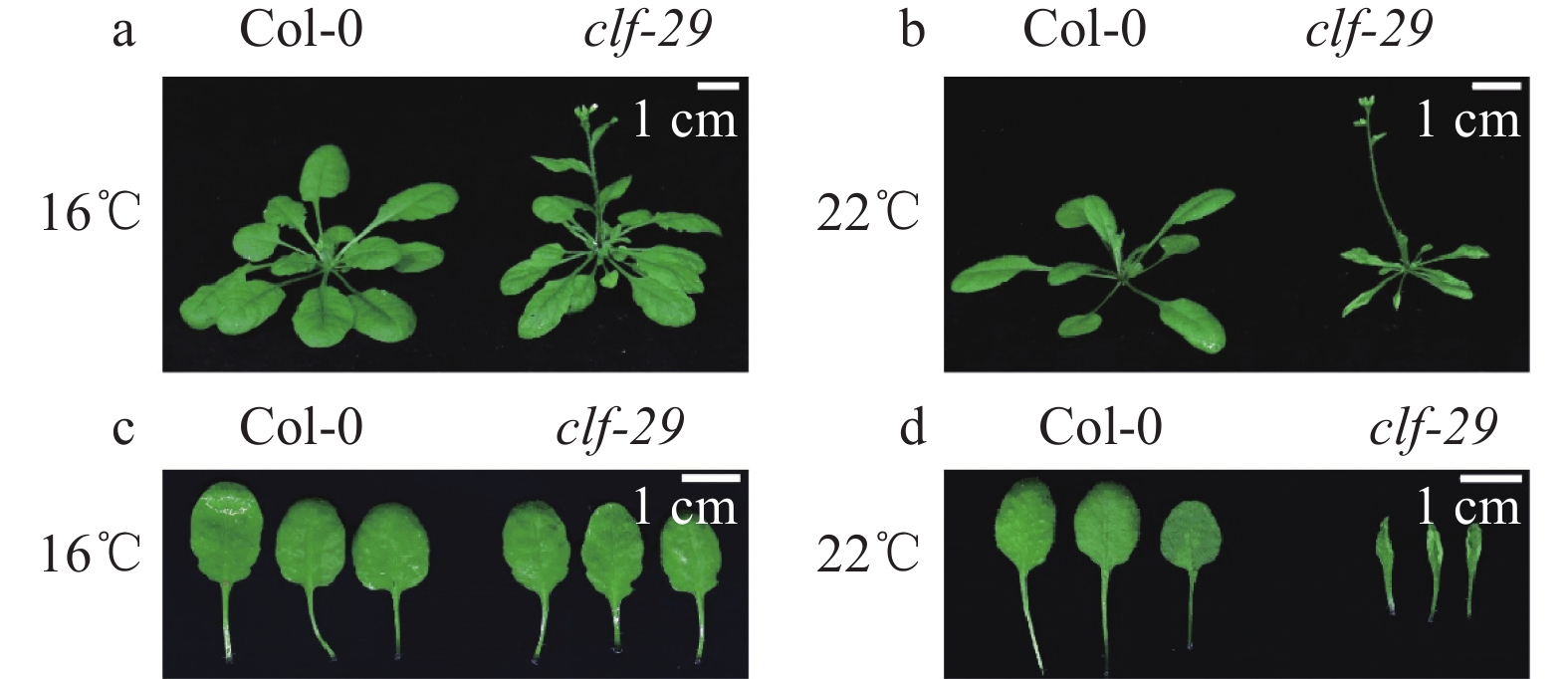
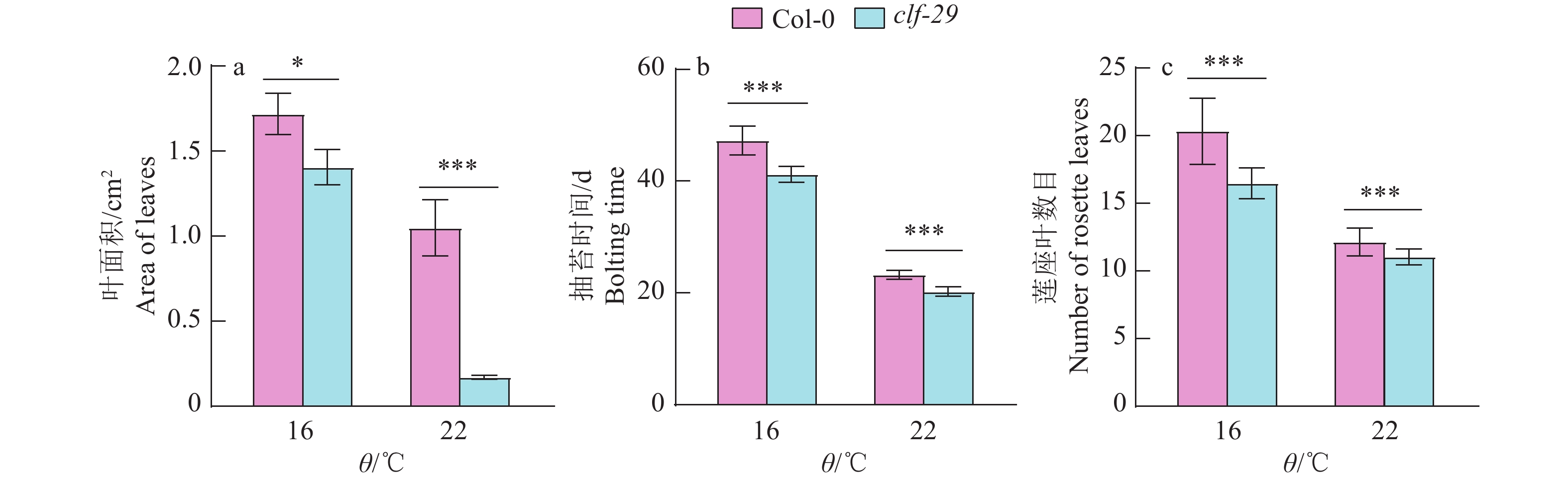
图 3 Col-0和clf-29在不同温度条件的表型统计
图a统计的叶片数量为3片,图b、c统计的植株数量为25株;“*”和“***”分别表示Col-0和clf-29在P < 0.05和P < 0.001水平差异显著(t检验)
Figure 3. Phenotypic statistics of Col-0 and clf-29 at different temperatures
The number of leaves counted in figure a is 3, the number of plants counted in figure b, c is 25; “*” and “***” indicate significant differences at P < 0.05 and P < 0.001 levels between Col-0 and clf-29 respectively (t test)
表 1 本研究用到的引物
Table 1 Primers used in this study
用途
Usage名称
Name序列
Sequence基因分型
Genotypingclf-29-BP 5′-ATTTTGCCGATTTCGGAAC-3′ clf-29-LP 5′-AAGAAACTTGCTAGTTCCGCC-3′ clf-29-RP 5′-GAGGCATTGACTTTGATTTGC-3′ RT-qPCR SUS1-F 5′-GGCTAGGCTTGATCGTGTCA-3′ SUS1-R 5′-GATCCACCTGAACTGACCGT-3′ LHCA1-F 5′-CAGTCCCGTGGGGTACTTTG-3′ LHCA1-R 5′-GCCGCCCGTTCTTGATCTC-3′ B1L-F 5′-AATCTCCGATGGACCGTTTGA-3′ B1L-R 5′-AGAGCTTTCTTAGCTCGCCG-3′ DIN10-F 5′-CGCTTTCTGATCTTGGAAATCGC-3′ DIN10-R 5′-ACACCGGTTAGAATCGTCCG-3′ ACTIN 2-F 5′-AGTGTTAGCTGCTGCCGCTGT-3′ ACTIN 2-R 5′-ACCAGCAAAACCAGCCTTCACCA-3′ 表 2 RNA-seq数据统计1)
Table 2 Statistical analyses of RNA-seq data
θ/ ℃ 样品
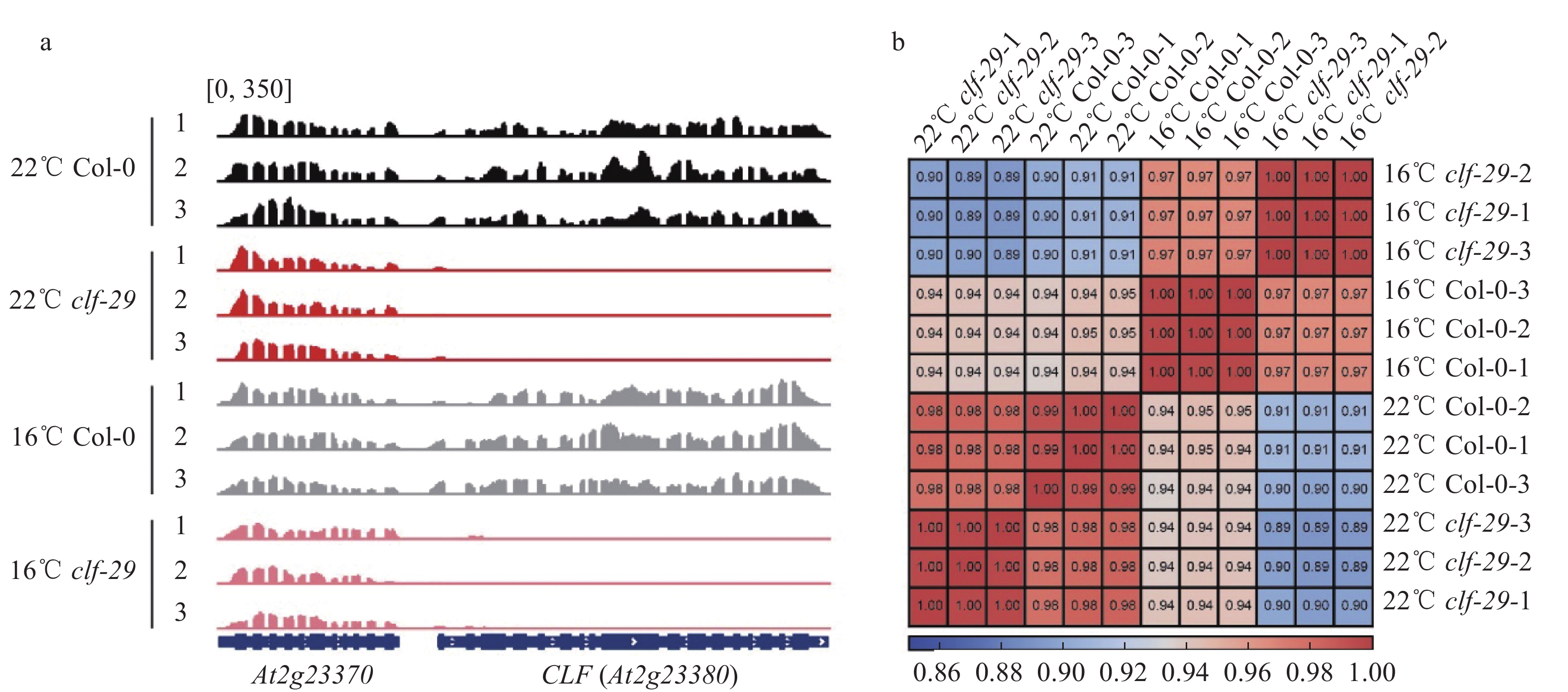
SampleNraw Ntrim ηtrim/% Nmap ηmap/% Nfilter ηfilter/% 22 Col-0-1 37 244 778 37 224 204 99.94 36 546 723 98.18 32 101 104 87.84 Col-0-2 22 235 624 22 206 546 99.87 21 582 542 97.19 19 117 943 88.58 Col-0-3 33 187 892 33 167 450 99.94 32 477 567 97.92 22 820 849 70.27 clf-29-1 34956528 34 928 744 99.92 34 052 033 97.49 30 609 312 89.89 clf-29-2 32 170 570 32 138 512 99.90 31 122 935 96.84 27 113 262 87.12 clf-29-3 33 342 874 33 311 222 99.91 32 488 435 97.53 29 058 947 89.44 16 Col-0-1 22 773 106 22 750 524 99.90 22 163 560 97.42 20 064 343 90.53 Col-0-2 28 556 576 28 521 012 99.88 27 642 565 96.92 24 617 366 89.06 Col-0-3 29 554 426 29 506 424 99.84 28 694 997 97.25 26 892 274 93.72 clf-29-1 31 924 456 31 886 896 99.88 30 662 439 96.16 28 160 281 91.84 clf-29-2 34 539 696 34 510 886 99.92 33 692 978 97.63 29 642 353 87.98 clf-29-3 30 517 278 30 473 504 99.86 29 717 761 97.52 25 571 998 86.05 1) Nraw:原始测序的reads数目;Ntrim、ηtrim:除去低质量碱基后的reads数目和对应的比例;Nmap、ηmap:比对到拟南芥基因组的reads数目和对应的比例;Nfilter、ηfilter:比对到拟南芥染色质上且高比对质量的reads数目和对应的比例
1) Nraw: The number of raw reads; Ntrim, ηtrim: The number and corresponding proportion of reads after removing the low quality bases; Nmap, ηmap: The number and corresponding proportion of reads that aligned on Arabidopsi genome; Nfilter, ηfilter: The number and corresponding proportion of reads that aligned on Arabidopsi with high quality -
[1] CHANG Y N, ZHU C, JIANG J, et al. Epigenetic regulation in plant abiotic stress responses[J]. Journal of Integrative Plant Biology, 2020, 62(5): 563-580. doi: 10.1111/jipb.12901
[2] LAMERS J, VAN DER MEER T, TESTERINK C. How plants sense and respond to stressful environments[J]. Plant Physiology, 2020, 182(4): 1624-1635. doi: 10.1104/pp.19.01464
[3] ASHAPKIN V V, KUTUEVA L I, ALEKSANDRUSHKINA N I, et al. Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses[J]. International Journal of Molecular Sciences, 2020, 21(20): 7457. doi: 10.3390/ijms21207457.
[4] GALLUSCI P, DAI Z, GENARD M, et al. Epigenetics for plant improvement: Current knowledge and modeling avenues[J]. Trends in Plant Science, 2017, 22(7): 610-623. doi: 10.1016/j.tplants.2017.04.009
[5] KASSIS J A, KENNISON J A, TAMKUN J W. Polycomb and trithorax group genes in Drosophila[J]. Genetics, 2017, 206(4): 1699-1725. doi: 10.1534/genetics.115.185116
[6] LEWIS E B. A gene complex controlling segmentation in Drosophila[J]. Nature, 1978, 276(5688): 565-570. doi: 10.1038/276565a0
[7] SCHUETTENGRUBER B, GANAPATHI M, LEBLANC B, et al. Functional anatomy of Polycomb and trithorax chromatin landscapes in Drosophila embryos[J]. PLoS Biology, 2009, 7(1): 146-163.
[8] SPARMANN A, VAN LOHUIZEN M. Polycomb silencers control cell fate, development and cancer[J]. Nature Reviews Cancer, 2006, 6(11): 846-856. doi: 10.1038/nrc1991
[9] PIEN S, GROSSNIKLAUS U. Polycomb group and trithorax group proteins in Arabidopsis[J]. Biochimica et Biophysica Acta: Gene Structure and Expression, 2007, 1769(5/6): 375-382.
[10] GOODRICH J, PUANGSOMLEE P, MARTIN M, et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis[J]. Nature, 1997, 386(6620): 44-51. doi: 10.1038/386044a0
[11] GROSSNIKLAUS U, VIELLE-CALZADA J P, HOEPPNER M A, et al. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis[J]. Science, 1998, 280(5362): 446-450. doi: 10.1126/science.280.5362.446
[12] CHANVIVATTANA Y, BISHOPP A, SCHUBERT D, et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis[J]. Development, 2004, 131(21): 5263-5276. doi: 10.1242/dev.01400
[13] XIAO J, WAGNER D. Polycomb repression in the regulation of growth and development in Arabidopsis[J]. Current Opinion in Plant Biology, 2015, 23: 15-24. doi: 10.1016/j.pbi.2014.10.003
[14] SHU J, CHEN C, THAPA R K, et al. Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings[J]. Plant Direct, 2019, 3(1): 100. doi: 10.1002/pld3.100.
[15] KIM G T, TSUKAYA H, UCHIMIYA H. The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana[J]. Planta, 1998, 206(2): 175-183. doi: 10.1007/s004250050389
[16] LAFOS M, KROLL P, HOHENSTATT M L, et al. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation[J]. PLoS Genetics, 2011, 7(4): e1002040. doi: 10.1371/journal.pgen.1002040
[17] LIU J, DENG S, WANG H, et al. CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis[J]. Plant Physiology, 2016, 171(1): 424-436. doi: 10.1104/pp.15.01335
[18] GU X, XU T, HE Y. A histone H3 lysine-27 methyltransferase complex represses lateral root formation in Arabidopsis thaliana[J]. Molecular Plant, 2014, 7(6): 977-988. doi: 10.1093/mp/ssu035
[19] DING Y, SHI Y, YANG S. Molecular regulation of plant responses to environmental temperatures[J]. Molecular Plant, 2020, 13(4): 544-564. doi: 10.1016/j.molp.2020.02.004
[20] KUMAR S V, WIGGE P A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis[J]. Cell, 2010, 140(1): 136-147. doi: 10.1016/j.cell.2009.11.006
[21] GIL K E, PARK C M. Thermal adaptation and plasticity of the plant circadian clock[J]. New Phytologist, 2019, 221(3): 1215-1229. doi: 10.1111/nph.15518
[22] QUINT M, DELKER C, FRANKLIN K A, et al. Molecular and genetic control of plant thermomorphogenesis[J]. Nature Plants, 2016, 2(1): 15190. doi: 10.1038/NPLANTS.2015.190.
[23] BLAZQUEZ M A, AHN J H, WEIGEL D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana[J]. Nature Genetics, 2003, 33(2): 168-171. doi: 10.1038/ng1085
[24] MARTIN M. Cutadapt removes adapter sequences from high-throughput sequencing reads[J]. EMBnet Journal, 2011, 17(1): 10-12. doi: 10.14806/ej.17.1.200
[25] KIM D, LANGMEAD B, SALZBERG S L. HISAT: A fast spliced aligner with low memory requirements[J]. Nature Methods, 2015, 12(4): 357-360. doi: 10.1038/nmeth.3317
[26] LI H, HANDSAKER B, WYSOKER A, et al. The sequence alignment/map format and SAMtools[J]. Bioinformatics, 2009, 25(16): 2078-2079. doi: 10.1093/bioinformatics/btp352
[27] LIAO Y, SMYTH G K, SHI W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930. doi: 10.1093/bioinformatics/btt656
[28] LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology, 2014, 15(12): 550. doi: 10.1186/s13059-014-0550-8.
[29] RAMIREZ F, RYAN D P, GRUNING B, et al. DeepTools2: A next generation web server for deep-sequencing data analysis[J]. Nucleic Acids Research, 2016, 44(W1): W160-W165. doi: 10.1093/nar/gkw257
[30] THORVALDSDOTTIR H, ROBINSON J T, MESIROV J P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration[J]. Briefings in Bioinformatics, 2013, 14(2): 178-192. doi: 10.1093/bib/bbs017
[31] YU G, WANG L G, HAN Y, et al. ClusterProfiler: An R package for comparing biological themes among gene clusters[J]. OMICS: A Journal of Integrative Biology, 2012, 16(5): 284-287. doi: 10.1089/omi.2011.0118
[32] JUNG C G, HWANG S G, PARK Y C, et al. Molecular characterization of the cold- and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations[J]. Journal of Plant Physiology, 2015, 176: 138-146. doi: 10.1016/j.jplph.2015.01.001
[33] CHEN T, CHEN J H, ZHANG W, et al. BYPASS1-LIKE, a DUF793 family protein, participates in freezing tolerance via the CBF pathway in Arabidopsis[J]. Frontiers in Plant Science, 2019, 10: 807. doi: 10.3389/fpls.2019.00807.
[34] BOUREAU L, HOW-KIT A, TEYSSIER E, et al. A CURLY LEAF homologue controls both vegetative and reproductive development of tomato plants[J]. Plant Molecular Biology, 2016, 90(4/5): 485-501.
[35] LUO M, PLATTEN D, CHAUDHURY A, et al. Expression, imprinting, and evolution of rice homologs of the polycomb group genes[J]. Molecular Plant, 2009, 2(4): 711-723. doi: 10.1093/mp/ssp036
[36] KWON C S, LEE D, CHOI G, et al. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis[J]. Plant Journal, 2009, 60(1): 112-121. doi: 10.1111/j.1365-313X.2009.03938.x
[37] RAMAKRISHNAN M, ZHANG Z, MULLASSERI S, et al. Epigenetic stress memory: A new approach to study cold and heat stress responses in plants[J]. Frontiers in Plant Science, 2022, 13: 1075279. doi: 10.3389/fpls.2022.1075279.
[38] TIAN Y, ZHENG H, ZHANG F, et al. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR[J]. Science Advances, 2019, 5(4): eaau7246. doi: 10.1126/sciadv.aau7246
[39] YANG H, BERRY S, OLSSON T S G, et al. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis[J]. Science, 2017, 357(6356): 1142-1145. doi: 10.1126/science.aan1121



 下载:
下载: