Cloning and functional verification of circadian clock gene MtTOC1a in Medicago truncatula
-
摘要:目的
分析蒺藜苜蓿Medicago truncatula生物钟基因MtTOC1a的蛋白结构,探究MtTOC1a在生物钟系统中的生物学功能,比较其与拟南芥Arabidopsis thaliana的AtTOC1功能相似性和差异性。
方法通过生物信息学分析,在全基因组范围内鉴定了TOC1在蒺藜苜蓿中的同源基因。构建MtTOC1a基因的表达载体,利用农杆菌介导法引入到拟南芥野生型Col、及相应的功能丧失突变体toc1-2中,进行遗传互补分析。
结果MtTOC1a和MtTOC1b均具有保守的功能结构域和三维结构。遗传分析表明,在早期光形态建成中,外源转化的MtTOC1a完全恢复了toc1-2的下胚轴伸长表型,但对toc1-2的提前开花表型没有显著影响。在引入CAB::LUC报告基因的株系中,外源转化MtTOC1a在连续光照下使短周期突变体toc1-2的近日节律周期延长,但仍不能完全恢复至野生型水平。
结论MtTOC1a和拟南芥AtTOC1的功能存在相似性,但在不同的下游调控途径中所扮演的角色存在差异。本研究结果为进一步探索MtTOC1a基因的功能,利用MtTOC1a基因改造苜蓿的重要性状提供了理论依据。
Abstract:ObjectiveThe goal of this study is to analyze the protein structure of Medicago truncatula MtTOC1a, explore the biological function of MtTOC1a in the circadian clock system, and compare its similarities and differences in function with its ortholog AtTOC1 in Arabidopsis thaliana.
MethodThe orthologous genes of TOC1 in Medicago were identified through bioinformatics analysis, the expression vector of MtTOC1a gene was constructed and introduced into Arabidopsis wild-type Col and the corresponding loss-of-function mutant toc1-2 by Agrobacterium mediated method for genetic complementation analysis. Both MtTOC1a and MtTOC1b have conserved functional domains and protein structures. The genetic analysis indicated that during early photomorphogenesis, exogenously transformed MtTOC1a fully restored the hypocotyl elongation phenotype of toc1-2, but had no significant effect on the premature flowering phenotype of toc1-2. In the CAB::LUC reporter lines, MtTOC1a lengthened the period of the short period mutant toc1-2 under continuous light conditions, yet the mutant could not fully recover to the wild-type level.
ConclusionMtTOC1a and AtTOC1 have similar functions, but their roles in drownstream pathways are still different. The results provide a theoretical basis for further exploring the function of MtTOC1a gene and using MtTOC1a gene to modify the important traits in Medicago.
-
Keywords:
- Medicago truncatula /
- Arabidopsis thaliana /
- MtTOC1a /
- Circadian clock /
- Circadian rhythm /
- Agronomic trait
-
-
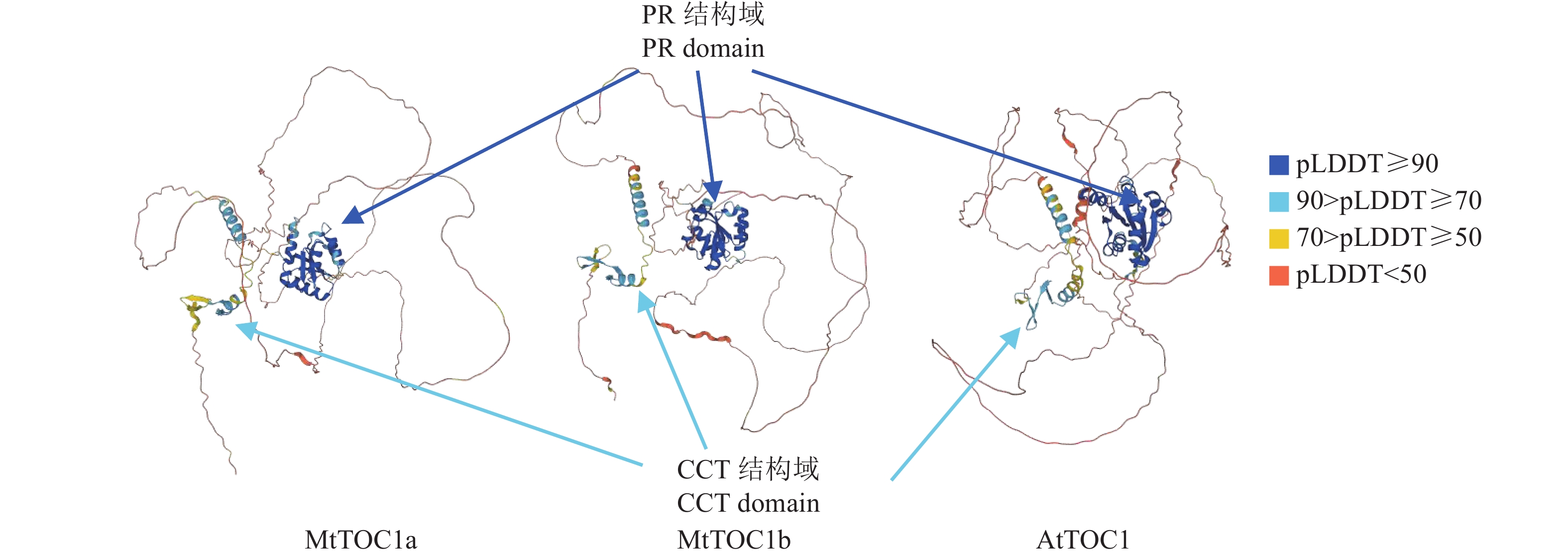
图 2 AlphaFold项目所预测的MtTOC1a、MtTOC1b和AtTOC1的蛋白三维结构
PR结构域与置信得分(pLDDT)≥90的深蓝色区域重合,呈多股α螺旋和β折叠构成的桶状结构;CCT结构域与90> pLDDT≥50的浅蓝色/黄色区域重合,由两条α螺旋构成类似剪刀状的结构
Figure 2. 3D protein structures of MtTOC1a, MtTOC1b and AtTOC1 predicted by the AlphaFold project
The PR domain overlaps with the dark blue region with a confidence score of (pLDDT) ≥90, showing a barrel-like structure composed of multiple strands of α helices and β folds; The CCT domain overlaps with the light blue/yellow region with 90> pLDDT≥50, forming a scissor-like structure with two α helices
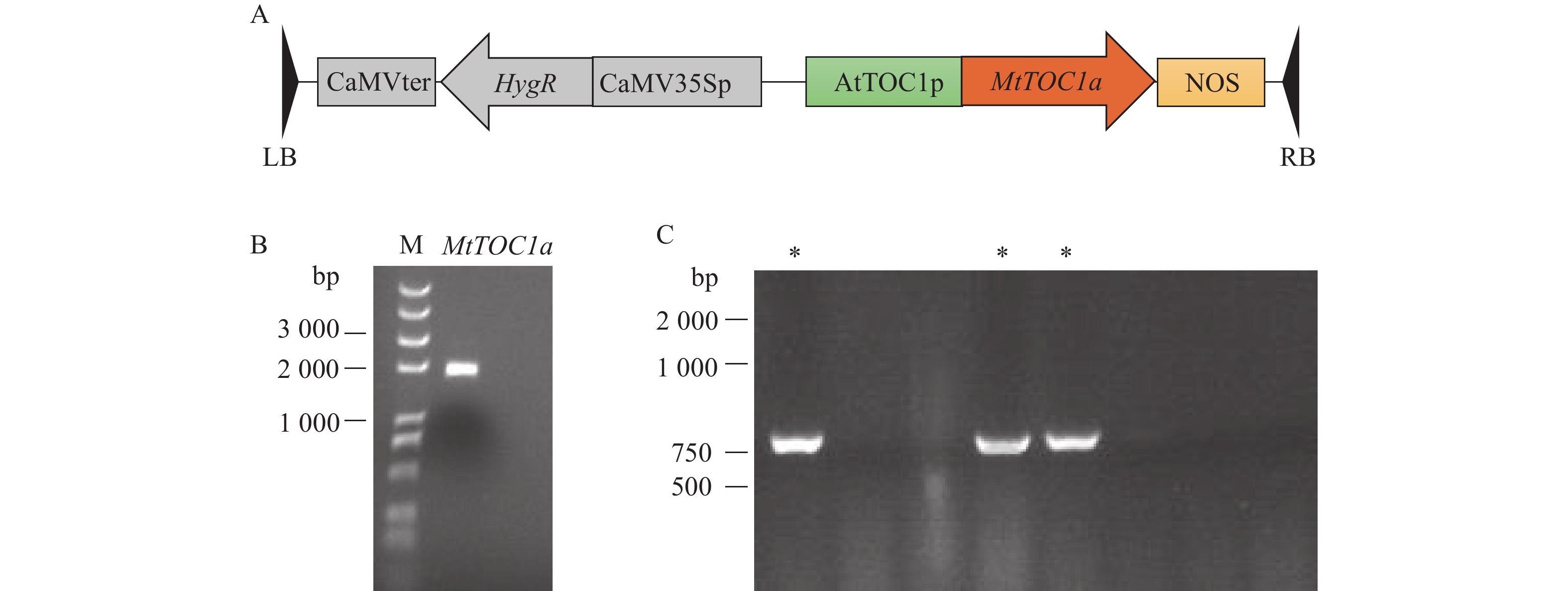
图 3 MtTOC1a表达载体的构建
A:MtTOC1a互补表达载体的T-DNA区域示意图;B:苜蓿MtTOC1a基因的克隆;C:MtTOC1a载体的菌落PCR结果,“*”表示阳性菌落
Figure 3. Construction of MtTOC1a expression vector
A: Schematic diagram of the T-DNA region of MtTOC1a expression vector; B: Cloning of MtTOC1a gene in Medicago; C: Colony PCR results of MtTOC1a expression vector, “*” indicates positive colony
图 5 MtTOC1a相关转基因植株的表型量化分析
A:苗龄7 d的下胚轴长度,n≥30;B:在短日照条件下植株抽薹时莲座叶的数量, n≥15;柱子上方的不同小写字母表示差异显著(P<0.05,LSD法)
Figure 5. Quantification analysis of the phenotypes of transgenic plants with MtTOC1a
A: The hypocotyl lengths of 7-day-old seedlings, n≥30; B: The number of rosette leaves during bolting of plants under short-day conditions, n≥15; Different lowercase letters on bars indicate significant differences (P<0.05, LSD test)
图 6 MtTOC1a相关转基因植株的近日节律周期分析
A:持续光照条件下的MtTOC1a相关转基因拟南芥植株生物发光节律,n≥16,图中所有植株均带有CAB::LUC荧光素酶报告基因,括号内表示相应植株的近日节律周期,浅灰色表示主观黑夜; B: A图中植株的近日节律周期和相对振幅误差的量化,相对振幅误差数值越小表示植株的节律性越强
Figure 6. Circadian rhythm analysis of transgenic plants with MtTOC1a
A: The bioluminescence rhythm of transgenic Arabidopsis plants with MtTOC1a under continuous light condition, n≥16, all plants in the figure carried a CAB:: LUC reporter gene, the daily rhythm cycle of corresponding plants was indicated in parentheses, light gray represents subjective night; B: Quantification of the circadian rhythm period and relative amplitude error of plants in A, and the smaller the relative amplitude error value, the stronger the rhythmicity of the plant
表 1 克隆载体的构建引物序列
Table 1 The primers used for cloning vector construction
基因
Gene引物名称
Primer name引物序列 (5′→3′)
Primer sequenceMtTOC1a MtTOC1a-F CTGATCATGGAGAGTGAAGGGTTTGATTTG MtTOC1a-R TTGCTCACCATAGCATCCCTCGGAGAGTAATCTC AtTOC1 AtTOC1pro-F CTCGGTACCCGGGGATCCGAGATCGCTCGGCTCAACAA AtTOC1pro-R TTCACTCTCCATGATCAGATTAACAACTAAACCCACACA -
[1] HARMER S L. The circadian system in higher plants[J]. Annual Review of Plant Biology, 2009, 60: 357-377. doi: 10.1146/annurev.arplant.043008.092054
[2] XU X, YUAN L, YANG X, et al. Circadian clock in plants: Linking timing to fitness[J]. Journal of Integrative Plant Biology, 2022, 64(4): 792-811. doi: 10.1111/jipb.13230
[3] MILLAR A J, CARRÉ I A, STRAYER C A, et al. Circadian clock mutants in Arabidopsis identified by luciferase imaging[J]. Science, 1995, 267(5201): 1161-1163. doi: 10.1126/science.7855595
[4] ALABADÍ D, OYAMA T, YANOVSKY M J, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock[J]. Science, 2001, 293(5531): 880-883. doi: 10.1126/science.1061320
[5] GENDRON J M, PRUNEDA-PAZ J L, DOHERTY C J, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(8): 3167-3172. doi: 10.1073/pnas.1200355109
[6] HUANG W, PÉREZ-GARCÍA P, POKHILKO A, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator[J]. Science, 2012, 336(6077): 75-79. doi: 10.1126/science.1219075
[7] LI N, ZHANG Y, HE Y, et al. Pseudo response regulators regulate photoperiodic hypocotyl growth by repressing PIF4/5 transcription[J]. Plant Physiology, 2020, 183(2): 686-699. doi: 10.1104/pp.19.01599
[8] DING Z, DOYLE M R, AMASINO R M, et al. A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation[J]. Genetics, 2007, 176(3): 1501-1510. doi: 10.1534/genetics.107.072769
[9] LEGNAIOLI T, CUEVAS J, MAS P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought[J]. The EMBO Journal, 2009, 28(23): 3745-3757. doi: 10.1038/emboj.2009.297
[10] VALIM H, DALTON H, JOO Y, et al. TOC1 in Nicotiana attenuata regulates efficient allocation of nitrogen to defense metabolites under herbivory stress[J]. New Phytologist, 2020, 228(4): 1227-1242. doi: 10.1111/nph.16784
[11] FUNG-UCEDA J, LEE K, SEO P J, et al. The circadian clock sets the time of DNA replication licensing to regulate growth in Arabidopsis[J]. Developmental Cell, 2018, 45(1): 101-113. doi: 10.1016/j.devcel.2018.02.022
[12] PECRIX Y, STATON S E, SALLET E, et al. Whole-genome landscape of Medicago truncatula symbiotic genes[J]. Nature Plants, 2018, 4(12): 1017-1025. doi: 10.1038/s41477-018-0286-7
[13] FARRÉ E M, LIU T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks[J]. Current Opinion in Plant Biology, 2013, 16(5): 621-629. doi: 10.1016/j.pbi.2013.06.015
[14] CORELLOU F, SCHWARTZ C, MOTTA J P, et al. Clocks in the green lineage: Comparative functional analysis of the circadian architecture of the Picoeukaryote ostreococcus[J]. The Plant Cell, 2009, 21(11): 3436-3449. doi: 10.1105/tpc.109.068825
[15] PETERSEN J, RREDHI A, SZYTTENHOLM J, et al. Evolution of circadian clocks along the green lineage[J]. Plant Physiology, 2022, 190(2): 924-937. doi: 10.1093/plphys/kiac141
[16] VALIM H F, MCGALE E, YON F, et al. The clock gene TOC1 in shoots, not roots, determines fitness of Nicotiana attenuata under drought[J]. Plant Physiology, 2019, 181(1): 305-318. doi: 10.1104/pp.19.00286
[17] BENDIX C, MARSHALL C M, HARMON F G. Circadian clock genes universally control key agricultural traits[J]. Molecular Plant, 2015, 8(8): 1135-1152. doi: 10.1016/j.molp.2015.03.003
[18] KONG Y, HAN L, LIU X, et al. The nodulation and nyctinastic leaf movement is orchestrated by clock gene LHY in Medicago truncatula[J]. Journal of Integrative Plant Biology, 2020, 62(12): 1880-1895. doi: 10.1111/jipb.12999
[19] WANG L, ZHOU A, WANG L, et al. Core clock component MtLUX controls shoot architecture through repression of MtTB1/MtTCP1A in Medicago truncatula[J]. The Crop Journal, 2023, 11(3): 723-732. doi: 10.1016/j.cj.2022.11.002
[20] KONG Y, ZHANG Y, LIU X, et al. The conserved and specific roles of the LUX ARRHYTHMO in circadian clock and nodulation[J]. International Journal of Molecular Sciences, 2022, 23(7): 3473. doi: 10.3390/ijms23073473



 下载:
下载: