Advances in molecular genetic mechanism of meiotic recombination and applications in crop breeding
-
摘要:
减数分裂是真核生物有性生殖产生染色体数目减半的单倍体配子所必需的生命过程。重组是减数分裂的核心事件之一,既增加了同源染色体间遗传信息的交换,又保证了其在减数分裂后期Ⅰ的正确分离。因此,减数分裂重组不仅增加了后代遗传多样性,还是作物遗传育种的基础。通过提高重组频率或改变其分布可以加速农作物育种进程,而降低或抑制重组可以固定杂种优势。近年来对植物减数分裂重组的分子遗传机制的研究取得了很大进展,包括重组的遗传和表观遗传调控机制,重组的遗传操控技术、固定杂交优势和染色体工程等方面。本文针对以上方面进行了全面的总结,这些内容不仅方便了读者对减数分裂重组的理论认知,还拓展了通过调控减数分裂重组操控生物育种的思路。
Abstract:Meiosis is essential for producing haploid gametes during sexual reproduction in most eukaryotes. Homologous recombination is one of the critical events of meiosis prophase I. It not only leads to the reshuffle of genetic information between homologs, but also ensures their proper segregation at anaphase I. Therefore, meiotic recombination is important to facilitate the genetic diversity and evolution among progeny, and also provides the theoretical basis for crop breeding. As expectedly, increasing the frequency of recombination or changing its distribution can benefit crop breeding, while reducing or inhibiting recombination can sustain heterosis. Over the past decades, numerous achievements have been made in understanding and utilizing meiotic recombination in plants, including mechanisms on genetic and epigenetic regulation of meiotic recombination, manipulation technologies on recombination, fixation of heterosis and chromosome engineering. In this review, we summarize the latest findings and technologies for regulating meiotic recombination, which will enable the readers to have an easy access to understand meiotic recombination, and also expand the idea of manipulating breeding through meiotic recombination.
-
-
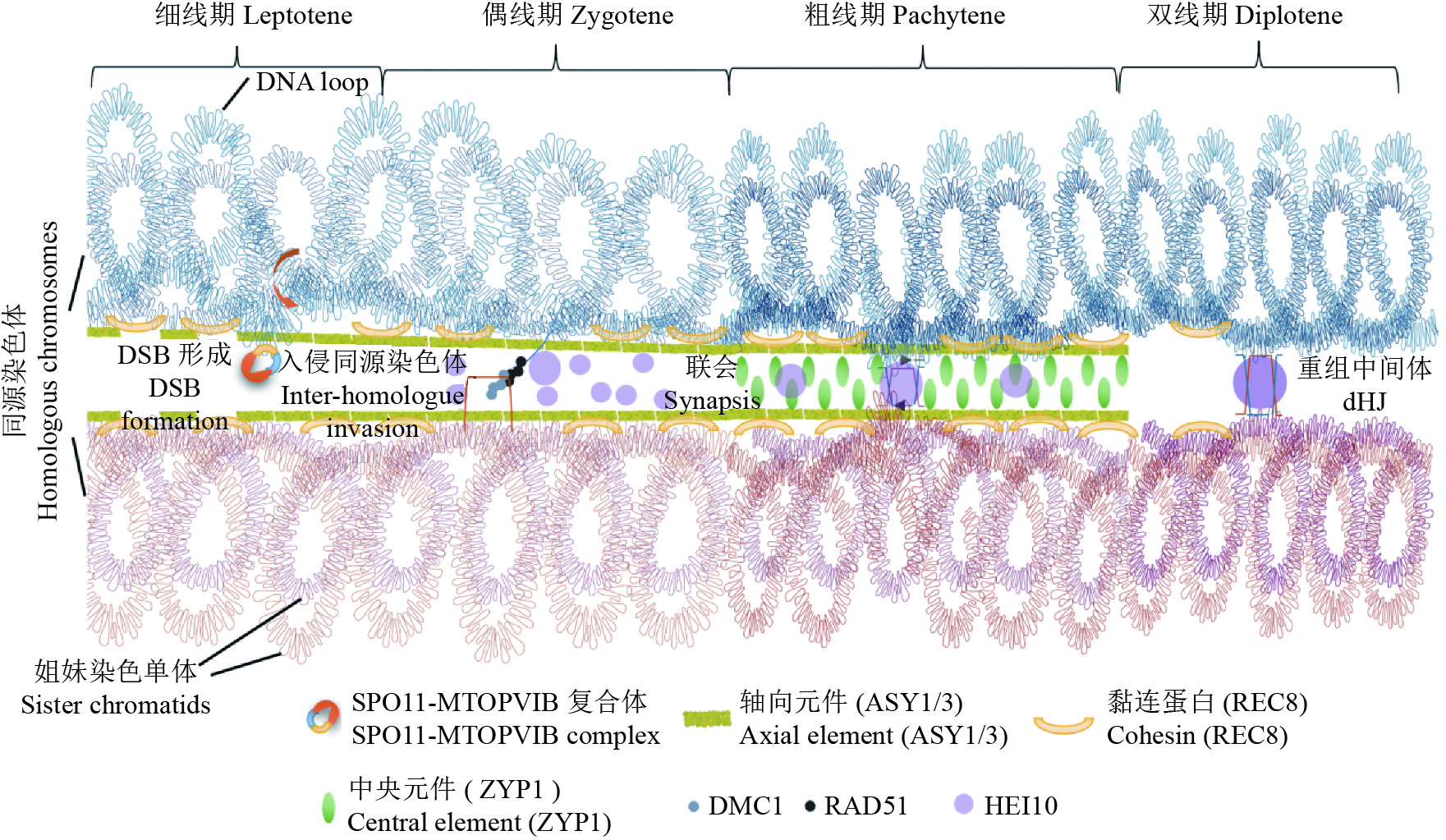
图 1 植物减数分裂染色体的牵回环–轴模型
减数分裂同源染色体的配对和联会是重组的重要保证。在减数分裂细线期,联会起始于轴向元件ASY1和ASY3在染色体上的加载,和黏连蛋白REC8一起最终形成一个线性的轴结构,使得姐妹染色单体沿着轴形成一个个DNA环。在细线期和偶线期转换时,DNA环会被牵拉到染色体轴上,由SPO11-MTOPVIB复合体介导双链断裂(DSBs)的产生。随后在偶线期,3′单链DNA会在RAD51-DMC1的介导下入侵到同源染色体间,形成D-loop,从而促进同源染色体配对。在粗线期,中央元件ZYP1加载到一对同源染色体轴中间,最终形成联会复合体的完整结构。在此时,大部分的DSB被修复,少量形成了重组中间体的结构。最后在双线期,联会复合体开始分解,同源染色体分离,只留下重组的区域形成交叉(Crossovers)
Figure 1. The model of tethered loop-axis of meiotic chromosomes in plant
The pairing and synapsis of meiotic homologous chromosomes are important for recombination. At leptotene, the synapsis initiates with the loading of axial elements ASY1 and ASY3 on the chromosome, which eventually forms a linear axis structure together with cohesin REC8. Sister chromosomes form DNA loops to array along the axis. During leptotene-diplotene transition, DNA loops are tethered onto the chromosome axis, and double-strand breaks (DSBs) are induced by SPO11-MTOPVIB complex. Subsequently, 3′ single-strand DNA end searches the homologous chromosome by RAD51-DMC1 to form a D-loop, thus promoting homologous chromosome pairing. At pachytene, the central element ZYP1 is loaded onto the centre of a pair of homologous chromosome axes, and finally forms the complete synaptonemal complex (SC) with axial elements. At this point, most DSBs are repaired, and a small amount forms recombination intermediates. At diplotene , following SC disassembly, homologs are separated except where crossovers have formed
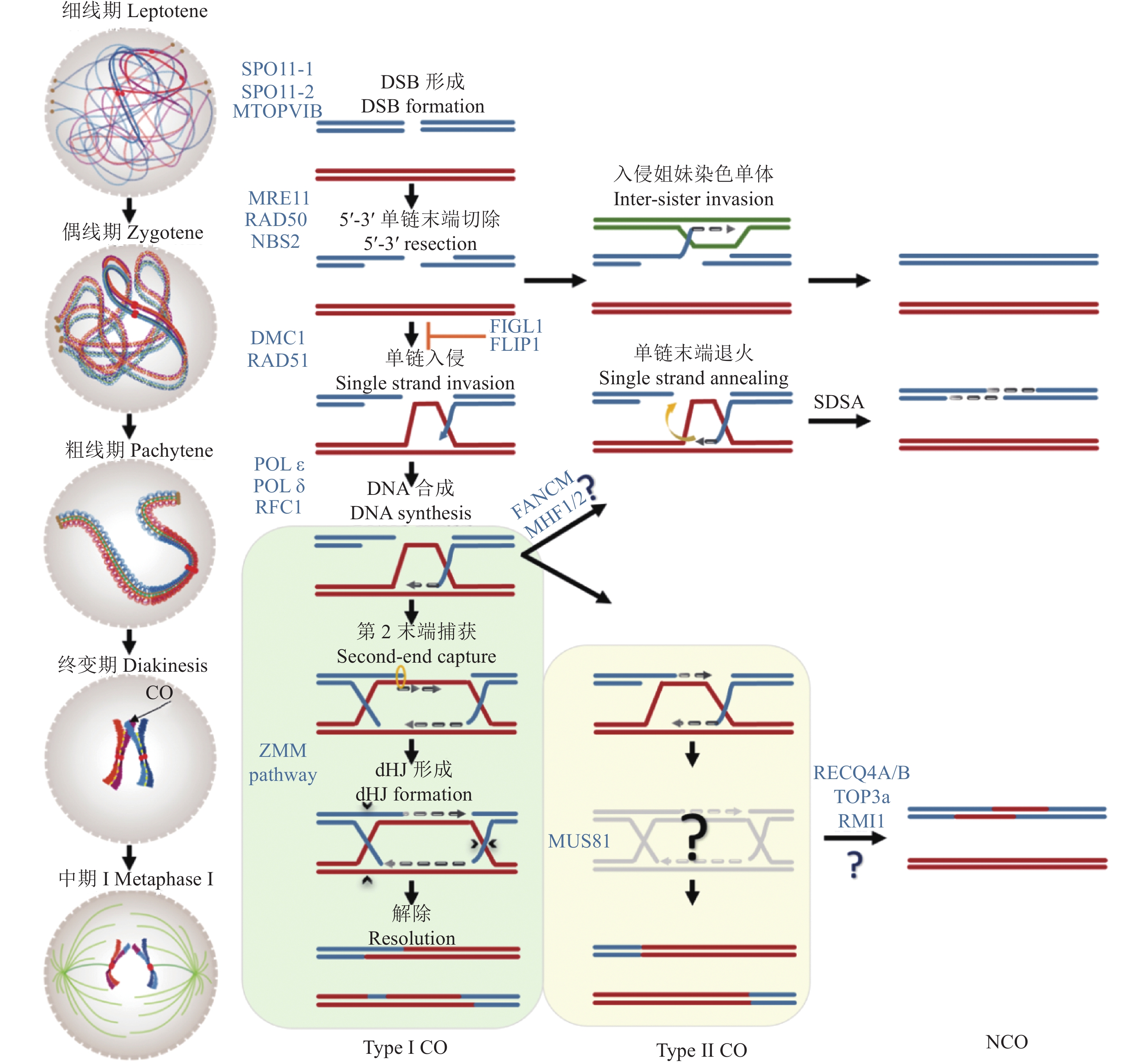
图 2 植物减数分裂重组修复途径模型
减数分裂的重组起始于双链断裂的形成,由SPO11-MTOPVIB复合体介导。双链断裂末端随后被切割处理形成3′单链尾巴。双链断裂修复可以选择以姐妹染色单体为模板,也可以在RAD51-DMC1的帮助下使3′单链末端入侵同源染色体形成置换环结构进行修复,后者形成减数分裂染色体重组。在以同源染色体为模板的修复中,DNA合成,第2链末端捕获和链接最终形成重组中间体的经典结构——双Holliday交叉,并最终解除形成干涉敏感型交换。同时,还存在着重组蛋白MUS81依赖的干涉不敏感型交换,但是目前植物中的重组中间体以及产物不太清楚。此外,在单链入侵后,当第2末端无法捕获时还存在一条合成依赖的链退火途径,最终也以姐妹染色单体为模板进行修复,并会产生非交换。在重组通路中,还存在着3条不同的重组抑制通路:FIGL1-FLIP、FANCM-MHF1/2和RECQ4A/B-TOP3α-RMI1,都参与抑制MUS81依赖的干涉不敏感型交换途径,并促进合成依赖的链退火
Figure 2. The model for meiotic recombination in plant
Meiotic recombination initiates with the formation of DSBs, which are mediated by the SPO11-MTOPVIB complex. DSB ends are cleaved to form 3′ single-strand tails. Subsequently, DSB can be repaired by selecting the sister chromatids as the template. Alternatively, 3′ single-strand ends invade the homolog to form a D-loop for repair, which is known as recombination. Following the repair progress, DNA synthesis, second strand end capture and ligation lead to the formation of double Holliday junctions (dHJs), which are the classical structure of the recombination intermediates and finally resolved as ZMM-dependent interference-sensitive crossovers (Type I COs). Meanwhile, MUS81-dependent pathway results in interference-insensitive crossovers (Type II CO) , but the recombination intermediates and their products in plants are not well understood. In addition, single-strand invasion can be processed by synthesis-dependent strand annealing pathway (SDSA), and chooses the sister chromatids as the template for repair to produce NCO. During meiotic recombination, there are also three different recombination inhibitory pathways, including FIGL1-FLIP, FANCM-MHF1/2 and RECQ4A/ B-Top3α-RMI1, which are involved in the inhibition of MUS81-dependent Type II CO and promote SDSA
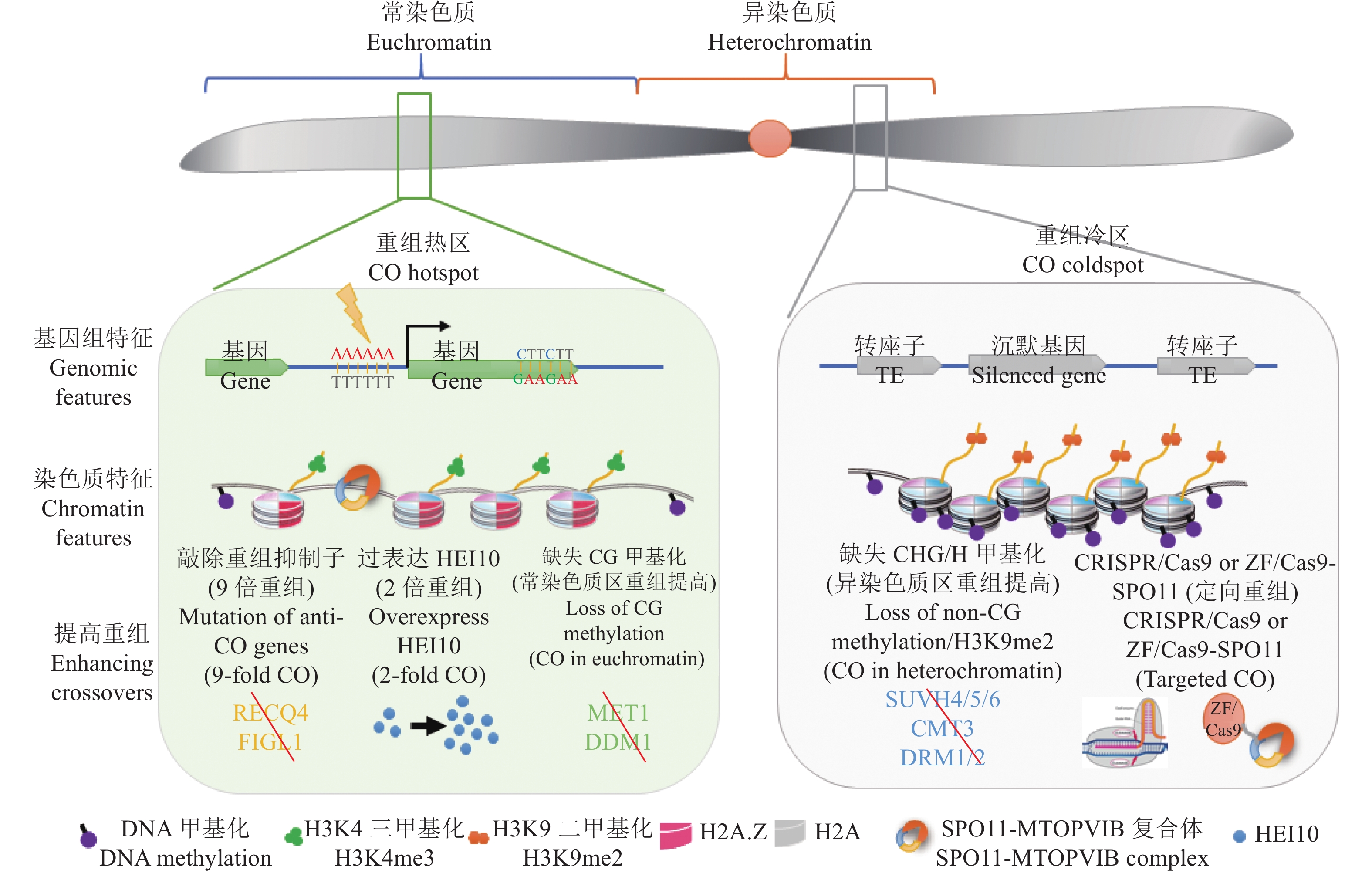
图 3 拟南芥减数分裂重组(CO)热点的基因组/染色质特征和调控模型
减数分裂重组在染色体上不是均匀分布的。通常倾向于发生在常染色质区,并且DSB通常发生于基因的转录起始位点或转录终止位点。重组热区伴随着低核小体密度、H2A.Z和H3K4me3的富集,并且含有AT-rich 和CTT 基序。而在异染色质区通常结构致密,TE和重复序列富集,伴随高非CG甲基化和H3K9me2,这些都是重组的抑制因素。在常染色质区提高减数分裂重组的方法有:突变重组抑制子、过表达重组酶HEI10和突变MET1和DDM1;而在异染色质区提高重组通常通过降低非CG甲基化或H3K9me2,另外还可以通过定向重组的方法提高目标区域重组
Figure 3. The model for regulation of Arabidopsis meiotic crossover hotspots by genomic and chromatin features
The distribution of meiotic recombination events is not uniformly along chromosomes. The crossovers (COs) generally tends to occur in euchromatin regions, and DSBs usually occur in transcription start sites (TSS) or transcription stop sites (TTS). Meiotic recombination hotspots display low nucleosome density, occupancy of H2A.Z and H3K4me3 enrichment with AT-rich and CTT motifs. Heterochromatin is highly compacted with lots of TE and repeat sequences accompanied by high non-CG methylation and H3K9me2, which are the inhibitors of recombination. The approaches for improving meiotic recombination frequency on euchromatin are as follows: Mutating anti-CO genes, overexpressing the recombinase HEI10 or mutating DNA methyltransferases MET1 and remodeler DDM1. Increasing COs on heterochromatin can be achieved by reducing non-CG methylation or H3K9me2, and targeted recombination
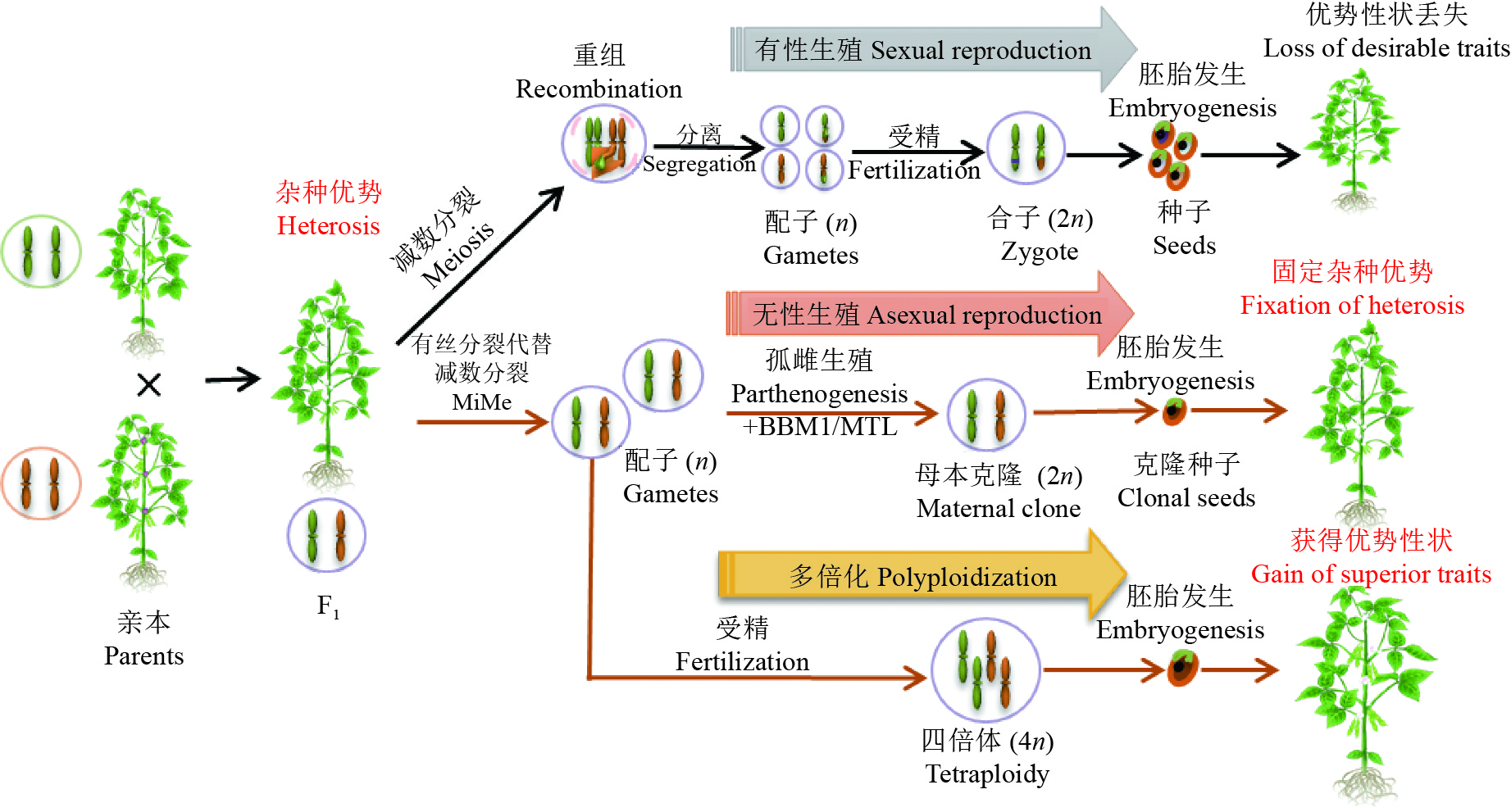
图 4 操纵减数分裂创新作物种质
植物杂交后代具有较亲本优越的表型或适应性称为杂种优势。在有性生殖过程中,F1代杂交植物经历减数分裂染色体重组和分离,导致配子间的等位基因再分配。而后植物双受精使雄配子与雌配子融合,由于减数分裂重组,在F1代中观察到的理想杂种优势性状通常在其后代中丢失。与此相反,无融合生殖依赖于未减数孢子分裂、孤雌生殖和不依赖受精的功能性胚乳的形成。在改良的植物无融合生殖中,通过对有丝分裂代替减数分裂(Mitosis instead of Meiosis, MiMe)和BBM1/MTL基因的遗传操作,分别诱导有丝分裂代替细胞分裂,产生二倍体配子,而后通过孤雌生殖实现种子克隆。克隆种子与F1杂交植株基因一致,可以在后代中保持杂种优势。此外,还可以通过MiMe实现后代多倍化,从而提高后代植物重组频率,整合并增强优良性状
Figure 4. Manipulating meiosis for crop improvement
Heterosis is a phenomenon among progenies of plants, which is known as that hybrid plants can have superior traits compared to their parents. In the process of sexual reproduction, F1 hybrid plants undergo meiotic recombination and chromosome segregation, leading to redistribution of alleles among gametes. Then plant double fertilization is the fusion of male gametes and female gametes. Due to meiotic recombination, the desirable traits observed in F1 hybrid plants are usually lost in their progenies. In contrast, apomixis relies on the apomeiosis, parthenogenesis and autonomous endosperm. In the improved apomixis of plant, MiMe induced by genetic manipulation produces diploid gametes and misexpression of BBM1/MTL in egg cell triggers parthenogenesis, thereby producing the clonal reproduction through seeds. The genome of cloned seeds is consistent with F1 hybrid plants, which can maintain heterosis in the progeny. In addition, diploid gametes can be achieved through MiMe to produce polyploid offspring, which may improve the frequency of recombination and enhance superior traits
-
[1] BLAT Y, PROTACIO R U, HUNTER N, et al. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation[J]. Cell, 2002, 111(6): 791-802. doi: 10.1016/S0092-8674(02)01167-4
[2] ARMSTRONG S J, CARYL A P, JONES G H, et al. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica [J]. Journal of Cell Science, 2002, 115(Pt 18): 3645-3655.
[3] FERDOUS M, HIGGINS J D, OSMAN K, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3[J]. PLoS Genetics, 2012, 8(2): e1002507. doi: 10.1371/journal.pgen.1002507.
[4] KEENEY S, GIROUX C N, KLECKNER N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family[J]. Cell, 1997, 88(3): 375-384. doi: 10.1016/S0092-8674(00)81876-0
[5] VRIELYNCK N, CHAMBON A, VEZON D, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination[J]. Science, 2016, 351(6276): 939-943. doi: 10.1126/science.aad5196
[6] SUGIMOTO-SHIRASU K, STACEY N J, CORSAR J, et al. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis[J]. Current Biology, 2002, 12(20): 1782-1786. doi: 10.1016/S0960-9822(02)01198-3
[7] STACEY N J, KUROMORI T, AZUMI Y, et al. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination[J]. The Plant Journal, 2006, 48(2): 206-216. doi: 10.1111/j.1365-313X.2006.02867.x
[8] NEALE M J, PAN J, KEENEY S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks[J]. Nature, 2005, 436(7053): 1053-1057. doi: 10.1038/nature03872
[9] DE MUYT A, PEREIRA L, VEZON D, et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana[J]. PLoS Genetics, 2009, 5(9): e1000654. doi: 10.1371/journal.pgen.1000654.
[10] JI J H, TANG D, SHEN Y, et al. P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(38): 10577-10582. doi: 10.1073/pnas.1607334113
[11] WANG Y X, COPENHAVER G P. Meiotic recombination: Mixing it up in plants[J]. Annual Review of Plant Biology, 2018, 69: 577-609. doi: 10.1146/annurev-arplant-042817-040431
[12] MIAO C, TANG D, ZHANG H, et al. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice[J]. The Plant Cell, 2013, 25(8): 2998-3009. doi: 10.1105/tpc.113.113175
[13] PANIZZA S, MENDOZA M A, BERLINGER M, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination[J]. Cell, 2011, 146(3): 372-383. doi: 10.1016/j.cell.2011.07.003
[14] LAMBING C, TOCK A J, TOPP S D, et al. Interacting genomic landscapes of REC8-cohesin, chromatin, and meiotic recombination in Arabidopsis[J]. The Plant Cell, 2020, 32(4): 1218-1239. doi: 10.1105/tpc.19.00866
[15] LAMBING C, KUO P C, TOCK A J, et al. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(24): 13647-13658. doi: 10.1073/pnas.1921055117
[16] LAM I, KEENEY S. Mechanism and regulation of meiotic recombination initiation[J]. Cold Spring Harbor Perspectives in Biology, 2014, 7(1): a016634. doi: 10.1101/cshperspect.a016634.
[17] UANSCHOU C, SIWIEC T, PEDROSA-HARAND A, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene[J]. The EMBO Journal, 2007, 26(24): 5061-5070. doi: 10.1038/sj.emboj.7601913
[18] OSMAN K, HIGGINS J D, SANCHEZ-MORAN E, et al. Pathways to meiotic recombination in Arabidopsis thaliana[J]. New Phytologist, 2011, 190(3): 523-544. doi: 10.1111/j.1469-8137.2011.03665.x
[19] WATERWORTH W M, ALTUN C, ARMSTRONG S J, et al. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants[J]. The Plant Journal, 2007, 52(1): 41-52. doi: 10.1111/j.1365-313X.2007.03220.x
[20] JI J H, TANG D, WANG M J, et al. MRE11 is required for homologous synapsis and DSB processing in rice meiosis[J]. Chromosoma, 2013, 122(5): 363-376. doi: 10.1007/s00412-013-0421-1
[21] JI J, TANG D, WANG K, et al. The role of OsCOM1 in homologous chromosome synapsis and recombination in rice meiosis [J] The Plant Journal, 2012, 72(1): 18-30.
[22] TSUBOUCHI H, OGAWA H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae[J]. Molecular Biology of the Cell, 2000, 11(7): 2221-2233. doi: 10.1091/mbc.11.7.2221
[23] HU Q, TANG D, WANG H J, et al. The exonuclease homolog OsRAD1 promotes accurate meiotic double-strand break repair by suppressing nonhomologous end joining[J]. Plant Physiology, 2016, 172(2): 1105-1116.
[24] CHE L X, WANG K J, TANG D, et al. OsHUS1 facilitates accurate meiotic recombination in rice[J]. PLoS Genetics, 2014, 10(6): e1004405. doi: 10.1371/journal.pgen.1004405.
[25] WOLD M S. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism[J]. Annual Review of Biochemistry, 1997, 66(1): 61-92. doi: 10.1146/annurev.biochem.66.1.61
[26] SHI B L, XUE J Y, YIN H, et al. Dual functions for the ssDNA-binding protein RPA in meiotic recombination[J]. PLoS Genetics, 2019, 15(2): e1007952. doi: 10.1371/journal.pgen.1007952.
[27] BROWN M S, BISHOP D K. DNA strand exchange and RecA homologs in meiosis[J]. Cold Spring Harbor Perspectives in Biology, 2014, 7(1): a016659. doi: 10.1101/cshperspect.a016659.
[28] AKLILU B B, SODERQUIST R S, CULLIGAN K M. Genetic analysis of the replication protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication[J]. Nucleic Acids Research, 2014, 42(5): 3104-3118. doi: 10.1093/nar/gkt1292
[29] ISHIBASHI T, KIMURA S, SAKAGUCHI K. A higher plant has three different types of RPA heterotrimeric complex[J]. The Journal of Biochemistry, 2006, 139(1): 99-104. doi: 10.1093/jb/mvj014
[30] MIAO Y J, SHI W Q, WANG H J, et al. Replication protein A large subunit (RPA1a) limits chiasma formation during rice meiosis[J]. Plant Physiology, 2021, 187(3): 1605-1618. doi: 10.1093/plphys/kiab365
[31] FERNANDES J B, DUHAMEL M, SEGUéLA -ARNAUD M, et al. FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination[J]. PLoS Genetics, 2018, 14(4): e1007317. doi: 10.1371/journal.pgen.1007317.
[32] DA INES O, ABE K, GOUBELY C, et al. Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana[J]. PLoS Genetics, 2012, 8(4): e1002636. doi: 10.1371/journal.pgen.1002636.
[33] DA INES O, DEGROOTE F, GOUBELY C, et al. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role[J]. PLoS Genetics, 2013, 9(9): e1003787. doi: 10.1371/journal.pgen.1003787.
[34] CLOUD V, CHAN Y L, GRUBB J, et al. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis[J]. Science, 2012, 337(6099): 1222-1225. doi: 10.1126/science.1219379
[35] SEELIGER K, DUKOWIC-SCHULZE S, WURZ-WILDERSINN R, et al. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana[J]. The New Phytologist, 2012, 193(2): 364-375. doi: 10.1111/j.1469-8137.2011.03947.x
[36] LIN Z, KONG H, NEI M, et al. Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(27): 10328-10333. doi: 10.1073/pnas.0604232103
[37] MERCIER R, MÉZARD C, JENCZEWSKI E, et al. The molecular biology of meiosis in plants[J]. Annual Review of Plant Biology, 2015, 66: 297-327. doi: 10.1146/annurev-arplant-050213-035923
[38] SU H, CHENG Z, HUANG J, et al. Arabidopsis RAD51, RAD51C and XRCC3 proteins form a complex and facilitate RAD51 localization on chromosomes for meiotic recombination[J]. PLoS Genetics, 2017, 13(5): e1006827. doi: 10.1371/journal.pgen.1006827.
[39] WANG Y, XIAO R, WANG H, et al. The Arabidopsis RAD51 paralogs RAD51B, RAD51D and XRCC2 play partially redundant roles in somatic DNA repair and gene regulation[J]. New Phytologist, 2014, 201(1): 292-304. doi: 10.1111/nph.12498
[40] DA INES O, DEGROOTE F, AMIARD S, et al. Effects of XRCC2 and RAD51B mutations on somatic and meiotic recombination in Arabidopsis thaliana[J]. The Plant Journal, 2013, 74(6): 959-970. doi: 10.1111/tpj.12182
[41] COLLINS I, NEWLON C S. Chromosomal DNA replication initiates at the same origins in meiosis and mitosis[J]. Molecular and Cellular Biology, 1994, 14(5): 3524-3534.
[42] SZOSTAK J W, ORR-WEAVER T L, ROTHSTEIN R J, et al. The double-strand-break repair model for recombination[J]. Cell, 1983, 33(1): 25-35. doi: 10.1016/0092-8674(83)90331-8
[43] TERASAWA M, OGAWA H, TSUKAMOTO Y, et al. Meiotic recombination-related DNA synthesis and its implications for cross-over and non-cross-over recombinant formation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(14): 5965-5970. doi: 10.1073/pnas.0611490104
[44] LU P L, HAN X W, QI J, et al. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis[J]. Genome Research, 2012, 22(3): 508-518. doi: 10.1101/gr.127522.111
[45] YANG H X, LU P L, WANG Y X, et al. The transcriptome landscape of Arabidopsis male meiocytes from high-throughput sequencing: The complexity and evolution of the meiotic process[J]. The Plant Journal, 2011, 65(4): 503-16. doi: 10.1111/j.1365-313X.2010.04439.x
[46] WANG Y X, CHENG Z H, HUANG J Y, et al. The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana[J]. PLoS Genetics, 2012, 8(11): e1003039. doi: 10.1371/journal.pgen.1003039.
[47] HUANG J Y, CHENG Z H, WANG C, et al. Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(40): 12534-12539. doi: 10.1073/pnas.1507165112
[48] HUANG J Y, COPENHAVER G P, MA H, et al. New insights into the role of DNA synthesis in meiotic recombination[J]. Science Bulletin, 2016, 61(16): 1260-1269. doi: 10.1007/s11434-016-1126-7
[49] WANG C, HUANG J Y, ZHANG J, et al. The largest subunit of DNA polymerase delta is required for normal formation of meiotic type I crossovers[J]. Plant Physiology, 2019, 179(2): 446-459. doi: 10.1104/pp.18.00861
[50] MCVEY M, KHODAVERDIAN V Y, MEYER D, et al. Eukaryotic DNA polymerases in homologous recombination[J]. Annual Review of Genetics, 2016, 50: 393-421. doi: 10.1146/annurev-genet-120215-035243
[51] MAGA G, STUCKI M, SPADARI S, et al. DNA polymerase switching: I: Replication factor C displaces DNA polymerase alpha prior to PCNA loading[J]. Journal of Molecular Biology, 2000, 295(4): 791-801. doi: 10.1006/jmbi.1999.3394
[52] DE MAAGD R A, LOONEN A, CHOUAREF J, et al. CRISPR/Cas inactivation of RECQ4 increases homeologous crossovers in an interspecific tomato hybrid[J]. Plant Biotechnology Journal, 2020, 18(3): 805-813. doi: 10.1111/pbi.13248
[53] MALOISEL L, BHARGAVA J, ROEDER G S. A role for DNA polymerase delta in gene conversion and crossing over during meiosis in Saccharomyces cerevisiae[J]. Genetics, 2004, 167(3): 1133-1142. doi: 10.1534/genetics.104.026260
[54] BISHOP D K, ZICKLER D. Early decision: Meiotic crossover interference prior to stable strand exchange and synapsis[J]. Cell, 2004, 117(1): 9-15. doi: 10.1016/S0092-8674(04)00297-1
[55] KURZBAUER M T, UANSCHOU C, CHEN D, et al. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis[J]. The Plant Cell, 2012, 24(5): 2058-2070. doi: 10.1105/tpc.112.098459
[56] REN L J, ZHAO T T, ZHAO Y Z, et al. The E3 ubiquitin ligase DESYNAPSIS1 regulates synapsis and recombination in rice meiosis[J]. Cell Reports, 2021, 37(5): 109941. doi: 10.1016/j.celrep.2021.109941.
[57] BENYAHYA F, NADAUD I, DA INES O, et al. SPO11.2 is essential for programmed double-strand break formation during meiosis in bread wheat (Triticum aestivum L. )[J]. The Plant Journal, 2020, 104(1): 30-43. doi: 10.1111/tpj.14903
[58] CRISMANI W, GIRARD C, FROGER N, et al. FANCM limits meiotic crossovers[J]. Science, 2012, 336(6088): 1588-1590. doi: 10.1126/science.1220381
[59] GIRARD C, CRISMANI W, FROGER N, et al. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers[J]. Nucleic Acids Research, 2014, 42(14): 9087-9095. doi: 10.1093/nar/gku614
[60] FERNANDES J B, SÉGUÉLA-ARNAUD M, LARCHEVÊQUE C, et al. Unleashing meiotic crossovers in hybrid plants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2431-2436. doi: 10.1073/pnas.1713078114
[61] MERCIER R, JOLIVET S, VEZON D, et al. Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not[J]. Current Biology, 2005, 15(8): 692-701. doi: 10.1016/j.cub.2005.02.056
[62] LYNN A, SOUCEK R, BÖRNER G V. ZMM proteins during meiosis: Crossover artists at work[J]. Chromosome Research, 2007, 15(5): 591-605. doi: 10.1007/s10577-007-1150-1
[63] MACAISNE N, VIGNARD J, MERCIER R. SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers [J]. Journal of Cell Science, 2011, 124(Pt 16): 2687-91.
[64] CHELYSHEVA L, VEZON D, CHAMBON A, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3[J]. PLoS Genetics, 2012, 8(7): e1002799. doi: 10.1371/journal.pgen.1002799.
[65] CHELYSHEVA L, GENDROT G, VEZON D, et al. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana[J]. PLoS Genetics, 2007, 3(5): e83. doi: 10.1371/journal.pgen.0030083.
[66] HIGGINS J D, ARMSTRONG S J H, FRANKLIN F C, et al. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis[J]. Genes & Development, 2004, 18(20): 2557-2570.
[67] HIGGINS J D, VIGNARD J, MERCIER R, et al. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis[J]. The Plant Journal, 2008, 55(1): 28-39. doi: 10.1111/j.1365-313X.2008.03470.x
[68] MAZINA O M, MAZIN A V, NAKAGAWA T, et al. Saccharomyces cerevisiae Mer3 helicase stimulates 3'-5' heteroduplex extension by Rad51: Implications for crossover control in meiotic recombination[J]. Cell, 2004, 117(1): 47-56. doi: 10.1016/S0092-8674(04)00294-6
[69] SNOWDEN T, ACHARYA S, BUTZ C, et al. hMSH4-hMSH5 recognizes holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes[J]. Molecular Cell, 2004, 15(3): 437-451. doi: 10.1016/j.molcel.2004.06.040
[70] DE MUYT A, PYATNITSKAYA A, ANDRÉANI J, et al. A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation[J]. Genes & Development, 2018, 32(3/4): 283-296.
[71] PYATNITSKAYA A, ANDREANI J, GUÉROIS R, et al. The Zip4 protein directly couples meiotic crossover formation to synaptonemal complex assembly[J]. Genes & Development, 2022, 36(1/2): 53-69.
[72] REYNOLDS A, QIAO H Y, YANG Y, et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis[J]. Nature Genetics, 2013, 45(3): 269-278. doi: 10.1038/ng.2541
[73] TOBY G G, GHERRABY W, COLEMAN T R, et al. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels[J]. Molecular and Cellular Biology, 2003, 23(6): 2109-2122. doi: 10.1128/MCB.23.6.2109-2122.2003
[74] ZIOLKOWSKI P A, UNDERWOOD C J, LAMBING C, et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination[J]. Genes & Development, 2017, 31(3): 306-317.
[75] MORGAN C, FOZARD J A, HARTLEY M, et al. Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis[J]. Nature Communications, 2021, 12(1): 4674. doi: 10.1038/s41467-021-24827-w.
[76] KURZBAUER M T, PRADILLO M, KERZENDORFER C, et al. Arabidopsis thaliana FANCD2 promotes meiotic crossover formation[J]. The Plant Cell, 2018, 30(2): 415-428. doi: 10.1105/tpc.17.00745
[77] MORGAN T H. Heredity and sex[Z] //Columbia University Lectures. The jessup lectures, New York: Columbia University Press, 1913.
[78] CAPILLA-PÉREZ L, DURAND S, HUREL A, et al. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(12): e2023613118. doi: 10.1073/pnas.2023613118.
[79] NONOMURA K I, NAKANO M, MURATA K, et al. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis[J]. Molecular Genetics and Genomics, 2004, 271(2): 121-129. doi: 10.1007/s00438-003-0934-z
[80] YUAN W Y, LI X W, CHANG Y X, et al. Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis[J]. The Plant Journal, 2009, 59(2): 303-315. doi: 10.1111/j.1365-313X.2009.03870.x
[81] FRANCE M G, ENDERLE J, RÖHRIG S, et al. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(14): e2021671118. doi: 10.1073/pnas.2021671118.
[82] POCHON G, HENRY I M, YANG C, et al. The Arabidopsis Hop1 homolog ASY1 mediates cross-over assurance and interference[EB/OL]. BioRxiv, 2022: 46164616 (2022-03-17) [2022-08-20]. https://doi.org/10.1101/2022.03.17.484635.
[83] DURAND S, LIAN Q, JING J, et al. Dual control of meiotic crossover patterning[EB/OL]. BioRxiv, 2022: 491364 (2022-05-11) [2022-08-20]. https://doi.org/10.1101/2022.05.11.491364.
[84] ROG O, KÖHLER S, DERNBURG A F. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors[J]. eLife, 2017, 6: e21455. doi: 10.7554/eLife.21455.
[85] ZHANG L Y, STAUFFER W, ZWICKER D, et al. Crossover patterning through kinase-regulated condensation and coarsening of recombination nodules[EB/OL]. BioRxiv, 2021: 457865 (2021-08-26) [2022-08-20]. https://doi.org/10.1101/2021.08.26.457865.
[86] MAYER K F X, WAUGH R, LANGRIDGE P, et al. A physical, genetic and functional sequence assembly of the barley genome[J]. Nature, 2012, 491(7426): 711-716. doi: 10.1038/nature11543
[87] APPELS R, EVERSOLE K, STEIN N, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome[J]. Science, 2018, 361(6403): eaar7191. doi: 10.1126/science.aar7191.
[88] LI X, LI L, YAN J B. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize[J]. Nature Communications, 2015, 6: 6648. doi: 10.1038/ncomms7648.
[89] CONSORTIUM T G. The tomato genome sequence provides insights into fleshy fruit evolution[J]. Nature, 2012, 485(7400): 635-641. doi: 10.1038/nature11119
[90] MCCONAUGHY S J. Recombination hotspots in soybean [Glycine max (L. ) Merr. ], in Agronomy and Horticulture Department[D]. Lincoln: University of Nebraska, 2022.
[91] SCHMUTZ J, CANNON S B, SCHLUETER J, et al. Genome sequence of the palaeopolyploid soybean[J]. Nature, 2010, 463(7278): 178-183. doi: 10.1038/nature08670
[92] BLARY A, JENCZEWSKI E. Manipulation of crossover frequency and distribution for plant breeding[J]. Theoretical and Applied Genetics, 2019, 132(3): 575-592. doi: 10.1007/s00122-018-3240-1
[93] HSU Y M, FALQUE M, MARTIN O C. Quantitative modelling of fine-scale variations in the Arabidopsis thaliana crossover landscape[J]. Quantitative Plant Biology, 2022, 3: e3. doi: 10.1017/qpb.2021.17.
[94] PRATTO F, BRICK K, CHENG G, et al. Meiotic recombination mirrors patterns of germline replication in mice and humans[J]. Cell, 2021, 184(16): 4251-4267. doi: 10.1016/j.cell.2021.06.025
[95] UNDERWOOD C J, CHOI K, LAMBING C, et al. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation[J]. Genome Research, 2018, 28(4): 519-531. doi: 10.1101/gr.227116.117
[96] CHOI K, ZHAO X H, TOCK A J, et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions[J]. Genome Research, 2018, 28(4): 532-546. doi: 10.1101/gr.225599.117
[97] LIAN Q C, SOLIER V, WALKEMEIER B, et al. The megabase-scale crossover landscape is largely independent of sequence divergence[J]. Nature Communications, 2022, 13(1): 3828. doi: 10.1038/s41467-022-31509-8.
[98] YELINA N E, LAMBING C, HARDCASTLE T J, et al. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis[J]. Genes & Development, 2015, 29(20): 2183-2202.
[99] CHOI K, ZHAO X, KELLY K A, et al. Arabidopsis meiotic crossover hot spots overlap with H2A. Z nucleosomes at gene promoters[J]. Nature Genetics, 2013, 45(11): 1327-1336. doi: 10.1038/ng.2766
[100] MARAND A P, JANSKY S H, ZHAO H N, et al. Meiotic crossovers are associated with open chromatin and enriched with Stowaway transposons in potato[J]. Genome Biology, 2017, 18: 203. doi: 10.1186/s13059-017-1326-8.
[101] KIANIAN P M A, WANG M, SIMONS K, et al. Highresolution crossover mapping reveals similarities and differences of male and female recombination in maize[J]. Nature Communications, 2018, 9: 2370. doi: 10.1038/s41467-018-04562-5.
[102] TOCK A J, HOLLAND D M, JIANG W, et al. Crossover-active regions of the wheat genome are distinguished by DMC1, the chromosome axis, H3K27me3, and signatures of adaptation[J]. Genome Research, 2021, 31(9): 1614-1628. doi: 10.1101/gr.273672.120
[103] YELINA N E, CHOI K, CHELYSHEVA L, et al. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants[J]. PLoS Genetics, 2012, 8(8): e1002844. doi: 10.1371/journal.pgen.1002844.
[104] PONT C, LEROY T, SEIDEL M, et al. Tracing the ancestry of modern bread wheats[J]. Nature Genetics, 2019, 51(5): 905-911. doi: 10.1038/s41588-019-0393-z
[105] ROWAN B A, HEAVENS D, FEUERBORN T R, et al. An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features[J]. Genetics, 2019, 213(3): 771-787. doi: 10.1534/genetics.119.302406
[106] MARTÍN A C, SHAW P, PHILLIPS D, et al. Licensing MLH1 sites for crossover during meiosis[J]. Nature Communications, 2014, 5: 4580. doi: 10.1038/ncomms5580.
[107] TAM S M, HAYS J B, CHETELAT R T. Effects of suppressing the DNA mismatch repair system on homeologous recombination in tomato[J]. Theoretical and Applied Genetics, 2011, 123(8): 1445-1458. doi: 10.1007/s00122-011-1679-4
[108] ZHANG L, PICKERING R, MURRAY B. Direct measurement of recombination frequency in interspecific hybrids between Hordeum vulgare and H. bulbosum using genomic in situ hybridization [J]. Heredity, 1999, 83 ( Pt 3): 304-309.
[109] BLACKWELL A R, DLUZEWSKA J, SZYMANSKA-LEJMAN M, et al. MSH2 shapes the meiotic crossover landscape in relation to interhomolog polymorphism in Arabidopsis[J]. The EMBO Journal, 2020, 39(21): e104858. doi: 10.15252/embj.2020104858.
[110] GOODWILLIE C, KALISZ S, ECKERT C G, The evolutionary Enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence [J]. Annual Review of Ecology, Evolution, and Systematics, 2005, 36(1): 47-79.
[111] MONNAHAN P, KOLÁŘ F, BADUEL P, et al. Pervasive population genomic consequences of genome duplication in Arabidopsis arenosa[J]. Nature Ecology & Evolution, 2019, 3(3): 457-468.
[112] BAUDAT F, BUARD J, GREY C, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice[J]. Science, 2010, 327(5967): 836-840. doi: 10.1126/science.1183439
[113] SMAGULOVA F, GREGORETTI I V, BRICK K, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots[J]. Nature, 2011, 472(7343): 375-378. doi: 10.1038/nature09869
[114] SHILO S, MELAMED-BESSUDO C, DORONE Y, et al. DNA crossover motifs associated with epigenetic modifications delineate open chromatin regions in Arabidopsis[J]. The Plant Cell, 2015, 27(9): 2427-2436. doi: 10.1105/tpc.15.00391
[115] LIU S Z, YEH C T, JI T M, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome[J]. PLoS Genetics, 2009, 5(11): e1000733. doi: 10.1371/journal.pgen.1000733.
[116] SAINTENAC C, FAURE S, REMAY A, et al. Variation in crossover rates across a 3-Mb contig of bread wheat (Triticum aestivum) reveals the presence of a meiotic recombination hotspot[J]. Chromosoma, 2011, 120(2): 185-198. doi: 10.1007/s00412-010-0302-9
[117] DEMIRCI S, VAN DIJK A D J, SANCHEZ PEREZ G, et al. Distribution, position and genomic characteristics of crossovers in tomato recombinant inbred lines derived from an interspecific cross between Solanum lycopersicum and Solanum pimpinellifolium[J]. The Plant Journal, 2017, 89(3): 554-564. doi: 10.1111/tpj.13406
[118] HENDERSON I R. Control of meiotic recombination frequency in plant genomes[J]. Current Opinion in Plant Biology, 2012, 15(5): 556-561. doi: 10.1016/j.pbi.2012.09.002
[119] KUO P, DA INES O, LAMBING C. Rewiring meiosis for crop improvement[J]. Frontiers in Plant Science, 2021, 12: 708948. doi: 10.3389/fpls.2021.708948.
[120] UNDERWOOD C J, MERCIER R. Engineering apomixis: Clonal seeds approaching the fields[J]. Annual Review of Plant Biology, 2022, 73: 201-225. doi: 10.1146/annurev-arplant-102720-013958
[121] BERCHOWITZ L E, FRANCIS K E, BEY A L, et al. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana[J]. PLoS Genetics, 2007, 3(8): e132. doi: 10.1371/journal.pgen.0030132.
[122] HARTUNG F, SUER S, PUCHTA H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(47): 18836-18841. doi: 10.1073/pnas.0705998104
[123] SÉGUÉLA-ARNAUD M, CHOINARD S, LARCHEVÊQUE C, et al. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains[J]. Nucleic Acids Research, 2017, 45(4): 1860-1871.
[124] SÉGUÉLA-ARNAUD M, CRISMANI W, LARCHEVÊQUE C, et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(15): 4713-4718. doi: 10.1073/pnas.1423107112
[125] GIRARD C, CHELYSHEVA L, CHOINARD S, et al. Correction: AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms[J]. PLoS Genetics, 2015, 11(9): e1005448. doi: 10.1371/journal.pgen.1005448.
[126] BLARY A, GONZALO A, EBER F, et al. FANCM limits meiotic crossovers in Brassica crops[J]. Frontiers in Plant Science, 2018, 9: 368. doi: 10.3389/fpls.2018.00368.
[127] DESJARDINS S D, SIMMONDS J, GUTERMAN I, et al. FANCM promotes class I interfering crossovers and suppresses class II non-interfering crossovers in wheat meiosis[J]. Nature Communications, 2022, 13(1): 3644. doi: 10.1038/s41467-022-31438-6.
[128] MIEULET D, AUBERT G, BRES C, et al. Unleashing meiotic crossovers in crops[J]. Nature Plants, 2018, 4(12): 1010-1016. doi: 10.1038/s41477-018-0311-x
[129] RAZ A, DAHAN-MEIR T, MELAMED-BESSUDO C, et al. Redistribution of meiotic crossovers along wheat chromosomes by virus-induced gene silencing[J]. Frontiers in Plant Science, 2021, 11: 635139. doi: 10.3389/fpls.2020.635139.
[130] LI X, YU M S, BOLAÑOS-VILLEGAS P, et al. Fanconi anemia ortholog FANCM regulates meiotic crossover distribution in plants[J]. Plant Physiology, 2021, 186(1): 344-360. doi: 10.1093/plphys/kiab061
[131] ARRIETA M, MACAULAY M, COLAS I, et al. An induced mutation in HvRECQL4 increases the overall recombination and restores fertility in a barley HvMLH3 mutant background[J]. Frontiers in Plant Science, 2021, 12: 706560. doi: 10.3389/fpls.2021.706560.
[132] DE MUYT A, ZHANG L R, PIOLOT T, et al. E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis[J]. Genes & Development, 2014, 28(10): 1111-1123.
[133] SERRA H, LAMBING C, GRIFFIN C H, et al. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(10): 2437-2442. doi: 10.1073/pnas.1713071115
[134] TAAGEN E, BOGDANOVE A J, SORRELLS M E. Counting on crossovers: Controlled recombination for plant breeding[J]. Trends in Plant Science, 2020, 25(5): 455-465. doi: 10.1016/j.tplants.2019.12.017
[135] MELAMED-BESSUDO C, LEVY A A. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): E981-E988. doi: 10.1073/pnas.1120742109.
[136] LIU C L, CAO Y W, HUA Y F, et al. Concurrent disruption of genetic interference and increase of genetic recombination frequency in hybrid rice using CRISPR/Cas9[J]. Frontiers in Plant Science, 2021, 12: 757152. doi: 10.3389/fpls.2021.757152.
[137] WANG K J, WANG C, LIU Q, et al. Increasing the genetic recombination frequency by partial loss of function of the synaptonemal complex in rice[J]. Molecular Plant, 2015, 8(8): 1295-1298. doi: 10.1016/j.molp.2015.04.011
[138] NAGESWARAN D C, KIM J, LAMBING C, et al. HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1 and restricts meiotic crossovers in Arabidopsis[J]. Nature Plants, 2021, 7(4): 452-467. doi: 10.1038/s41477-021-00889-y
[139] HOCHHOLDINGER F, BALDAUF J A. Heterosis in plants[J]. Current Biology, 2018, 28(18): R1089-R1092. doi: 10.1016/j.cub.2018.06.041
[140] SPILLANE C, CURTIS M D, GROSSNIKLAUS U. Apomixis technology development: Virgin births in farmers’ fields?[J]. Nature Biotechnology, 2004, 22(6): 687-691. doi: 10.1038/nbt976
[141] WANG C, LIU Q, SHEN Y, et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes[J]. Nature Biotechnology, 2019, 37(3): 283-286. doi: 10.1038/s41587-018-0003-0
[142] MARIMUTHU M P A, JOLIVET S, RAVI M, et al. Synthetic clonal reproduction through seeds[J]. Science, 2011, 331(6019): 876. doi: 10.1126/science.1199682.
[143] D'ERFURTH I, JOLIVET S, FROGER N, et al. Turning meiosis into mitosis[J]. PLoS Biology, 2009, 7(6): e1000124. doi: 10.1371/journal.pbio.1000124.
[144] MIEULET D, JOLIVET S, RIVARD M, et al. Turning rice meiosis into mitosis[J]. Cell Research, 2016, 26(11): 1242-1254. doi: 10.1038/cr.2016.117
[145] CROMER L, HEYMAN J, TOUATI S, et al. OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM[J]. PLoS Genetics, 2012, 8(7): e1002865. doi: 10.1371/journal.pgen.1002865.
[146] RAVI M, MARIMUTHU M P A, SIDDIQI I. Gamete formation without meiosis in Arabidopsis[J]. Nature, 2008, 451(7182): 1121-1124.
[147] HSU P D, LANDER E S, ZHANG F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6): 1262-1278. doi: 10.1016/j.cell.2014.05.010
[148] RÖNSPIES M, DORN A, SCHINDELE P, et al. CRISPR-Cas-mediated chromosome engineering for crop improvement and synthetic biology[J]. Nature Plants, 2021, 7(5): 566-573. doi: 10.1038/s41477-021-00910-4
[149] SARNO R, VICQ Y, UEMATSU N, et al. Programming sites of meiotic crossovers using Spo11 fusion proteins[J]. Nucleic Acids Research, 2017, 45(19): e164. doi: 10.1093/nar/gkx739.
[150] YELINA N E, HOLLAND D, GONZALEZ-JORGE S, et al. Coexpression of MEIOTIC-TOPOISOMERASE VIB-dCas9 with guide RNAs specific to a recombination hotspot is insufficient to increase crossover frequency in Arabidopsis[J]. G3 (Genes Genomes Genetics), 2022, 12(7): jkac105. doi: 10.1093/g3journal/jkac105.
[151] MANCERA E, BOURGON R, BROZZI A, et al. High-resolution mapping of meiotic crossovers and non-crossovers in yeast[J]. Nature, 2008, 454(7203): 479-485. doi: 10.1038/nature07135
[152] WANG Y Z, VAN RENGS W M J, ZAIDAN M W A M, et al. Meiosis in crops: From genes to genomes[J]. Journal of Experimental Botany, 2021, 72(18): 6091-6109. doi: 10.1093/jxb/erab217
[153] FILLER HAYUT S, MELAMED BESSUDO C, LEVY A A. Targeted recombination between homologous chromosomes for precise breeding in tomato[J]. Nature Communications, 2017, 8: 15605. doi: 10.1038/ncomms15605.
[154] KOURANOV A, ARMSTRONG C, SHRAWAT A, et al. Demonstration of targeted crossovers in hybrid maize using CRISPR technology[J]. Communications Biology, 2022, 5(1): 53. doi: 10.1038/s42003-022-03004-9.
[155] BEYING N, SCHMIDT C, PACHER M, et al. CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis[J]. Nature Plants, 2020, 6(6): 638-645. doi: 10.1038/s41477-020-0663-x
[156] SCHMIDT C, FRANSZ P, RÖNSPIES M, et al. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering[J]. Nature Communications, 2020, 11(1): 4418. doi: 10.1038/s41467-020-18277-z.
[157] SCHWARTZ C, LENDERTS B, FEIGENBUTZ L, et al. CRISPR-Cas9-mediated 75.5-Mb inversion in maize[J]. Nature Plants, 2020, 6(12): 1427-1431. doi: 10.1038/s41477-020-00817-6
[158] GALLEGO-BARTOLOMÉ J, GARDINER J, LIU W L, et al. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(9): E2125-E2134. doi: 10.1073/pnas.1716945115.
[159] GALLEGO-BARTOLOMÉ J, LIU W L, KUO P H, et al. Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis[J]. Cell, 2019, 176(5): 1068-1082.e19. doi: 10.1016/j.cell.2019.01.029
[160] DIRKS R, VAN DUN K, DE SNOO C B, et al. Reverse breeding: A novel breeding approach based on engineered meiosis[J]. Plant Biotechnology Journal, 2009, 7(9): 837-845. doi: 10.1111/j.1467-7652.2009.00450.x
[161] WIJNKER E, DEURHOF L, VAN DE BELT J, et al. Hybrid recreation by reverse breeding in Arabidopsis thaliana[J]. Nature Protocols, 2014, 9(4): 761-772. doi: 10.1038/nprot.2014.049
[162] WIJNKER E, VAN DUN K, DE SNOO C B, et al. Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant[J]. Nature Genetics, 2012, 44(4): 467-470. doi: 10.1038/ng.2203
[163] CALVO‐BALTANÁS V, WIJNEN C L, YANG C, et al. Meiotic crossover reduction by virus‐induced gene silencing enables the efficient generation of chromosome substitution lines and reverse breeding in Arabidopsis thaliana[J]. The Plant Journal, 2020, 104(5): 1437-1452. doi: 10.1111/tpj.14990



 下载:
下载: