Research status and prospect on bacterial 3-oxoacyl-ACP-reductase
-
摘要:
3–酮脂酰ACP还原酶属于短链醇脱氢酶/还原酶(SDR)超家族蛋白,参与细菌脂肪酸及相关物质合成,负责催化3–酮脂酰ACP还原为3–羟脂酰ACP。3–酮脂酰ACP还原酶在细菌中广泛存在,且氨基酸序列较为保守,然而其生物学功能却展现出多样性。本文对近年来细菌3–酮脂酰ACP还原酶的结构特性、生物学功能和抑制剂等方面的研究进展进行综述,为加深对3–酮脂酰ACP还原酶的理解和抗菌药物的开发提供有益的借鉴。
-
关键词:
- 3–酮脂酰ACP还原酶 /
- II型脂肪酸合成系统 /
- 抑制剂
Abstract:3-oxoacyl-ACP-reductase, a member of short chain dehydrogenase/reductase(SDR) super family, participates in the biothsythesis of bacterial fatty acid and derivatives by catalyzing the reduction of 3-oxoacyl-ACP to 3-hydroxyacyl-ACP. 3-oxoacyl-ACP-reductase is ubiquitously exsit in bacterial and highly conversed in amino acid sequence, however, it has diverse biological functions. In this review, we summerize the advance of structures, biological functions and inhibitors of 3-oxoacyl-ACP-reductase in recent years. This paper will provide useful references for further understanding of 3-oxoacyl-ACP-reductase and antibacterial drug design.
-
Keywords:
- 3-oxoacyl-ACP-reductase /
- Fatty acid synthesis system II /
- Inhibitor
-
脂肪酸是细胞生物主要的组成成分之一,是细胞膜磷脂、三酰甘油酯(脂肪)和糖脂的重要组分[1]。按碳链是否有支链可分为直链脂肪酸和支链脂肪酸[2];按照主碳链的碳原子数目,可以分为偶数脂肪酸和奇数脂肪酸,其中细胞膜的主要成分多为偶数脂肪酸[3];按照含不饱和碳碳双键的数目,可以分为饱和脂肪酸、单不饱和脂肪酸和多不饱和脂肪酸,生物体中单不饱和脂肪酸居多[4],按照双键构型,不饱和脂肪酸又可分为顺式脂肪酸和反式脂肪酸,生物体的不饱和脂肪酸主要为顺式不饱和脂肪酸[5]。
细菌通过调节脂肪酸的种类和组成,调节细胞膜的流动性,适应不同的生长环境[6-7]。此外脂肪酸还可以通过β–氧化途径为细菌提供大量的能量,是一种重要的储能物质[8-9]。
细菌能够从头合成脂肪酸,除了为膜磷脂和糖脂的合成提供底物外,还可为硫辛酸、生物素、鼠李糖脂、聚羟基脂肪酸(PHA/PHB)、类脂A、N–脂酰高丝氨酸内酯类(AHLs)和可扩散信号分子(Diffusible signal factor, DSF)等的合成提供前体[3, 10-14]。
目前对细菌脂肪酸合成系统已经进行了深入的研究,对一些关键酶的催化机理、生物学功能及其多样性也有众多的综合性论述,但鲜见3–酮脂酰ACP还原酶(3-oxoacyl-ACP-reductase,OAR)的系统性总结。本文对细菌3–酮脂酰ACP还原酶的研究进行系统性总结,以便为今后的科学研究提供参考。
1. 脂肪酸合成系统
细胞生物有2种类型的脂肪酸合成系统。哺乳动物、真菌以及分枝杆菌采用I型脂肪酸合成系统(Fatty acid synthesis I, FAS I),特点为脂肪酸的合成由1个多结构域的复杂脂肪酸合酶催化,其中每个结构域催化1个特定的生化反应,而脂酰基团由酰基载体蛋白(Acyl carrier protein,ACP)结构域携带,在各结构域间传递脂酰基团[15-16]。FAS I的最终产物一般为棕榈酸。另外,该系统不合成不饱和脂肪酸[17]。
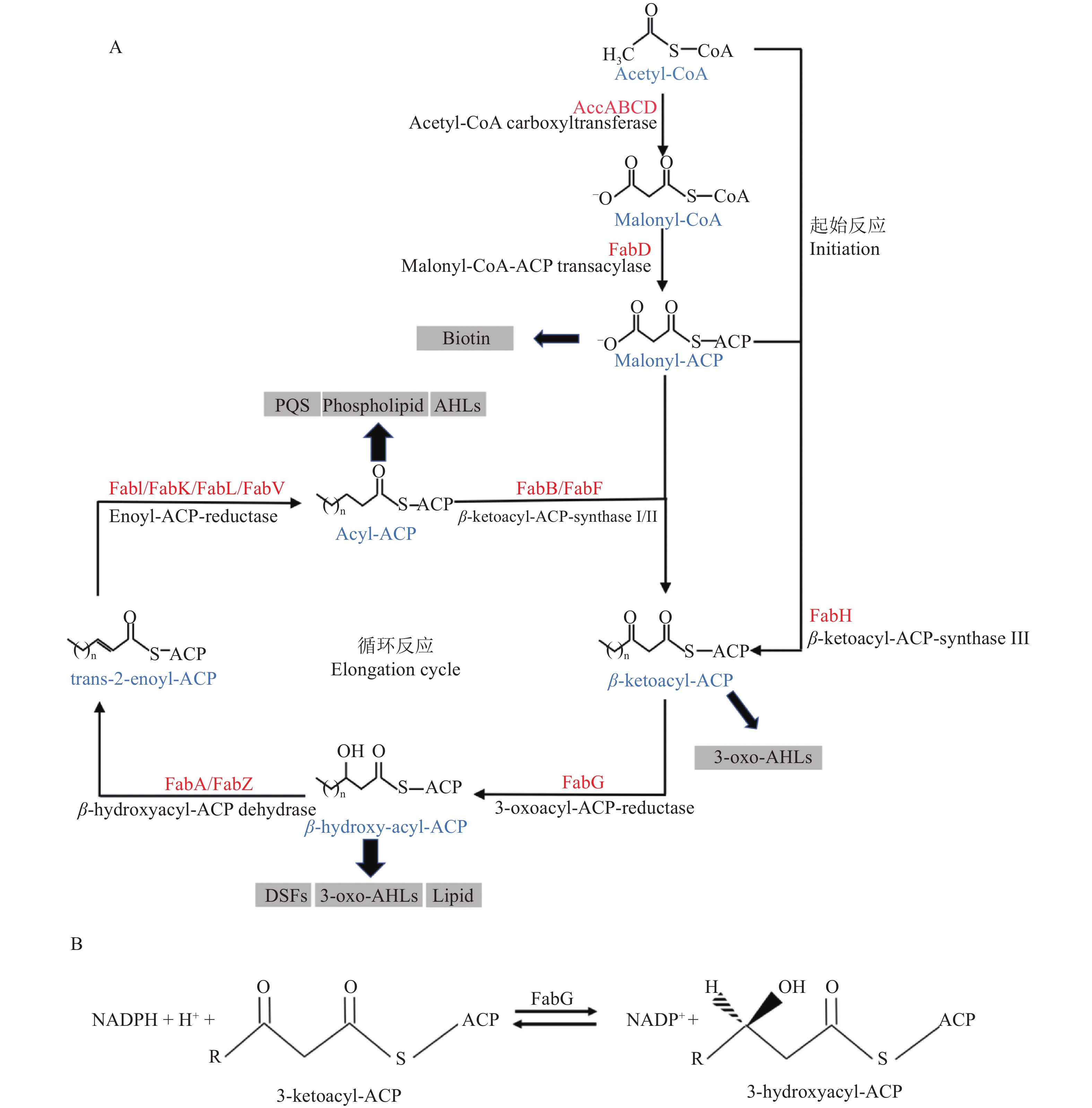
细菌和植物采用II型脂肪酸合成系统(Fatty acid synthesis II, FAS II)合成脂肪酸(图1A)[18]。FAS II的每一步反应均由独立的酶催化完成,脂酰基团由酰基载体蛋白ACP携带,可产生各种类型的脂酰ACP,为不同的最终产物提供前体[19-20]。同时,FAS II也可产生顺−3烯脂酰ACP,合成不饱和脂肪酸[5, 21-22]。因此,虽然I型和II型脂肪酸合成系统的生化反应相似,但参与催化的酶在结构和组成上有很大差异,这使得细菌脂肪酸合成途径成为开发新型抗菌药物的热门靶点[23-24]。
FAS II可分为起始反应和循环反应2部分(图1A)[18]。以大肠埃希菌Escherichia coli脂肪酸合成途径为例,起始反应的底物为乙酰–CoA,由乙酰辅酶A羧化酶(AccABCD)将其转化为丙二酸单酰–CoA,接着在丙二酸单酰辅酶A:ACP酰基转移酶(FabD)的催化下生产丙二酸单酰ACP,完成脂肪酸合成的起始反应[25]。循环反应的第1次循环由3–酮脂酰ACP合成酶III(FabH)将丙二酸单酰ACP和乙酰–CoA缩合,生成3–酮基丁酰ACP开始[26];接下来由3–酮脂酰ACP还原酶(FabG)催化,将3–酮基丁酰ACP还原为3–羟基丁酰ACP;然后由3–羟脂酰ACP脱水酶(FabA或FabZ)催化3–羟基丁酰ACP生成反–2–丁烯酰ACP[27-28];最后由烯脂酰ACP还原酶(FabI)催化反–2–丁烯酰ACP,生成丁酰ACP,至此完成第1次循环[29-30]。接下来的循环反应,由3–酮脂酰ACP合成酶I或II(FabB或FabF)催化脂酰ACP与丙二酸单酰ACP缩合开始,之后经FabG还原,FabA或FabZ脱水和FabI再次还原完成[31-32]。每一轮循环增加2个碳原子,直到合成棕榈酰ACP(C16–ACP)或硬脂酰ACP(C18–ACP)[33]。在大肠埃希菌中不饱和脂肪酸也可由FAS II酶催化合成。当碳链延伸到反–2–癸烯酰ACP时,双功能酶FabA行使其癸烯酰ACP异构酶的功能将反–2–癸烯酰ACP异构为顺–3–癸烯酰ACP,顺3–癸烯酰ACP不能被FabI催化,而是被FabB缩合进入下个循环反应,最终保留双键形成棕榈油酰ACP(C16:1)或异油酰ACP(C18:1)[34-35]。
2. 细菌脂肪酸合成中的3–酮脂酰ACP还原酶
细菌脂肪酸合成反应中,3–酮脂酰ACP还原酶(OAR)催化3–酮基脂酰ACP还原为3–羟基脂酰ACP。OAR是氧化还原酶,以NADPH为辅因子,极少数的OAR也可以利用NADH[23,36-37]。与参与细菌脂肪酸合成的其他酶不同,细菌OAR较为保守,目前已经报道的参与细菌脂肪酸合成的OAR均为大肠埃希菌FabG的同源蛋白,且编码基因均处在脂肪酸合成簇中。
大肠埃希菌FabG是首个报道的OAR,也是研究最为透彻的OAR。fabG基因位于脂肪酸合成基因簇plsX-fabH-fabD-fabG-acpP-fabF中。Zhang等[38]证明,fabG与上游的fabD基因共转录,在fabD与fabG的间隔序列中插入转录终止元件可使fabG无法转录。此外,转录终止元件的插入对大肠埃希菌是致死的,也证明了fabG是必需基因[39]。Lai等[40]构建了大肠埃希菌fabG的温度敏感突变菌株CL104。CL104在30 ℃时能够正常生长,在42 ℃时,由于FabG蛋白稳定性降低,菌体不能生长,这再次证明FabG是大肠埃希菌唯一参与脂肪酸合成的OAR。
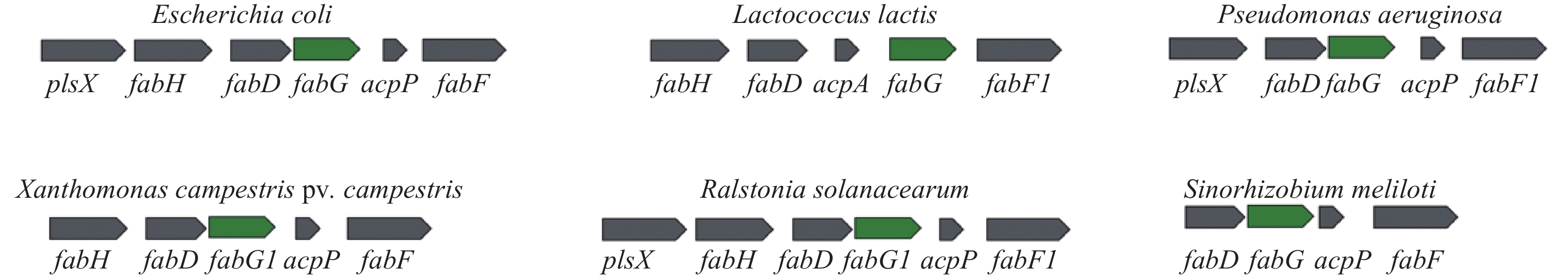
类似的脂肪酸合成基因簇在细菌中广泛存在。王海洪课题组先后对乳酸乳球菌Lactococcus lactis、茄科雷尔氏菌Ralstonia solanacearum、苜蓿中华根瘤菌Sinorhizobium meliloti、野油菜黄单胞菌Xanthomonas campestris pv. campestris和铜绿假单胞菌Pseudomonas aeruginosa脂肪酸合成基因簇中的fabG基因开展研究,结果证明这些fabG均为必需基因,且参与脂肪酸合成(图2)[41-45]。
除脂肪酸合成基因簇中的fabG被注释为OAR编码基因外,细菌基因组还有多个基因被注释为OAR编码基因。例如乳酸乳球菌基因组中有2个OAR编码基因:fabG1位于脂肪酸合成基因簇,fabG2所处的基因簇也与脂肪酸合成相关。但是研究表明只有fabG1编码的蛋白参与脂肪酸合成。茄科雷尔氏菌基因组包含1条染色体和1个巨质粒,生物信息学分析至少有4个基因被注释为OAR编码基因。fabG1(RSc1052)、fabG3(RSc0435)、fabG4(RSc0536)位于染色体上,而fabG2(RSp0359)位于巨质粒上。Feng等[42]研究表明,fabG2虽然能被敲除,突变菌株的脂肪酸组成显著改变,3–羟基脂肪酸合成量减少,而不饱和脂肪酸合成量增加,但是发现只有fabG1是必需基因,不能被敲除。在野油菜黄单胞菌基因组有4个被标注为OAR家族的编码基因:fabG1(XCC1018)、fabG2(XCC0416)、fabG3(XCC4003)和fabG4(XCC0384)。研究显示FabG1是一个典型的OAR,与大肠埃希菌FabG的序列一致性达到69.1%。FabG1在体内和体外均能够将3–酮脂酰ACP还原为3–羟脂酰ACP,fabG1是必需基因,只有在过表达其他OAR时才能将其敲除[44]。同样,铜绿假单胞菌中有12个基因被标注为OAR家族基因,然而只有位于脂肪酸合成基因簇的PA2967(fabG)参与脂肪酸合成[45];Cukier等[46]也通过构建条件突变菌株证明fabG是铜绿假单胞菌的必需基因。
2.1 细菌3–酮脂酰ACP还原酶结构特点
细菌脂肪酸合成中的OAR属于短链醇脱氢酶/还原酶(Short chain dehydrogenase/reductase, SDR)超家族[47]。SDR超家族是最大的蛋白质超家族之一,至少有160 000的蛋白属于此家族[48]。根据催化底物和关键活性残基的不同,SDR超家族可分为7个大类,464个家族。SDR超家族蛋白一级结构的序列一致性仅为15%~30%,但在序列和结构上有一些共同的特征。SDR超家族蛋白具有典型的Rossmann折叠结构,中心为6~7个β折叠,两侧各有3~4个α螺旋[49];催化活性残基为Tyr、Lys、Ser和Asn;辅因子为NADH或NADPH,辅因子结合位点位于N端[50]。与中链醇脱氢酶/还原酶(Medium chain dehydrogenase/reductase, MDR)超家族蛋白催化需要金属离子作为辅因子不同,SDR超家族蛋白催化不需要金属离子[51]。
OAR单体含有1个典型的Rossmann折叠结构,核心为7~8个β链构成的扭曲、平行的β折叠, 8个α螺旋依附于两侧。OAR的辅酶结合位点Gly-Xaa3-Gly-Xaa-Gly位于N端,中部含有Ser-Xaa12-Tyr-Xaa3-Lys催化基序。以上信息表明,OAR的序列和结构符合SDR超家族蛋白典型特征,属SDR家族中的129C亚家族[48]。
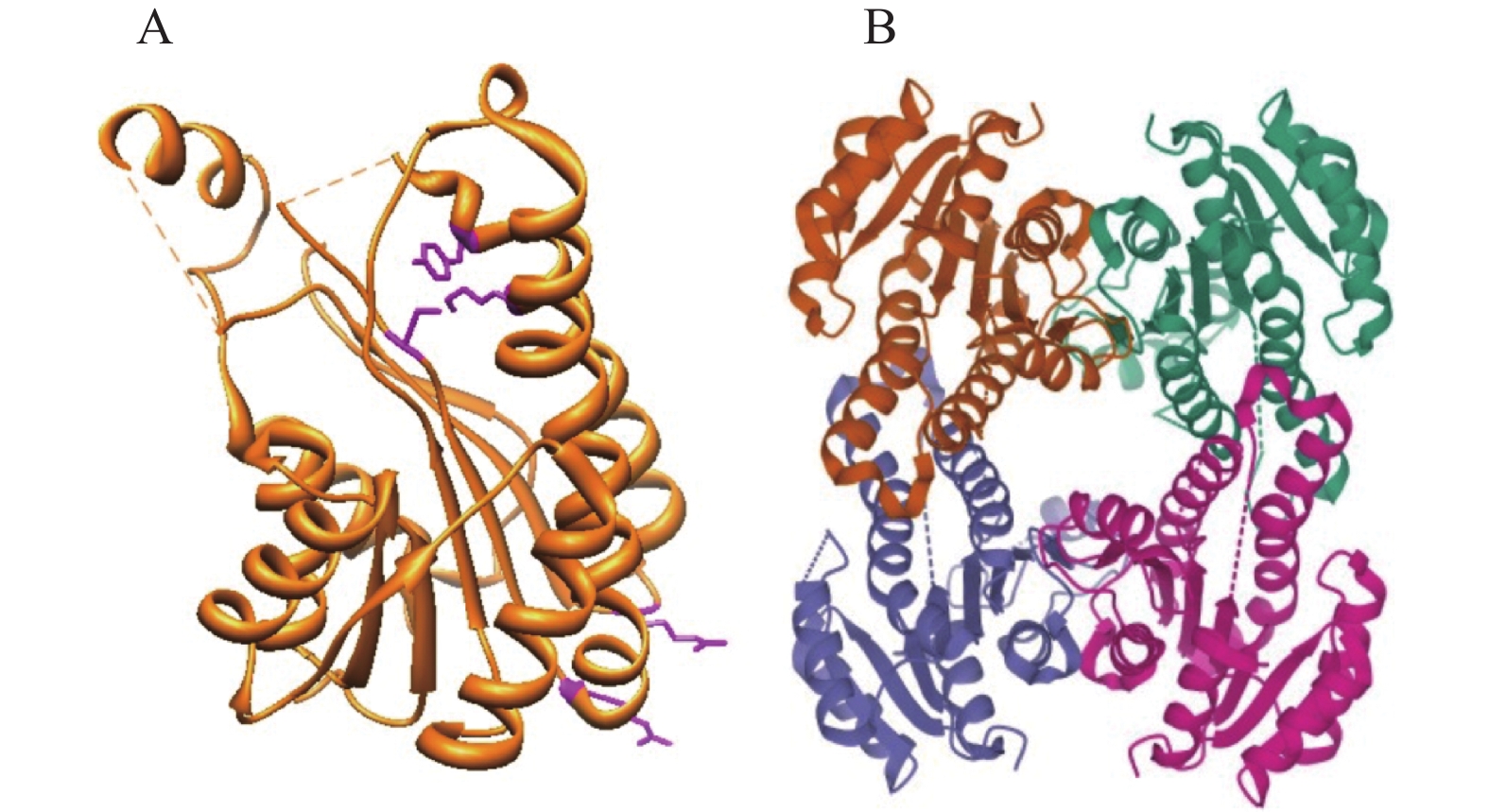
科学家们已经对大肠埃希菌、鼠伤寒沙门氏菌Salmonella enterica serovar Typhimurium、炭疽杆菌Bacillus anthracis、金黄色葡萄球菌Staphylococcus aureus、肺炎克雷伯菌Klebsiella pneumoniae、鲍曼不动杆菌Acinetobacter baumannii、结核分枝杆菌Mycobacterium tuberculosis等超过20个细菌的OAR进行了结构分析,所有的OAR都有着类似的三级结构[50, 52-56]。OAR一般为D2对称性的四聚体,即2个单体首先形成二聚体,2个二聚体再形成对称的四聚体。OAR结合NADPH后,形成3–酮基脂酰ACP-OAR-NADPH的复合体,OAR的构象发生改变,以容纳底物和辅因子,并传递电子[57]。
大肠埃希菌FabG是研究最为透彻的OAR,OAR的众多关键活性位点均是在FabG中报道的。Price等[54]首先解析了大肠埃希菌FabG的蛋白结构,FabG是典型的四聚体结构,4个单体形成2个二聚体界面,进而形成四聚体。FabG单体含有1个典型的Rossmann折叠结构,核心为7个β链构成的扭曲、平行的β折叠,通过8个α螺旋依附于亚基的两侧。2个单体的α4,α5螺旋和α4′,α5′螺旋形成疏水口袋,容纳3–酮基脂酰ACP的脂酰链。2个单体的β7折叠和α6/α7螺旋形成亲水的口袋,在结合NADPH及后续的构象改变中起着非常重要的作用(图3)。
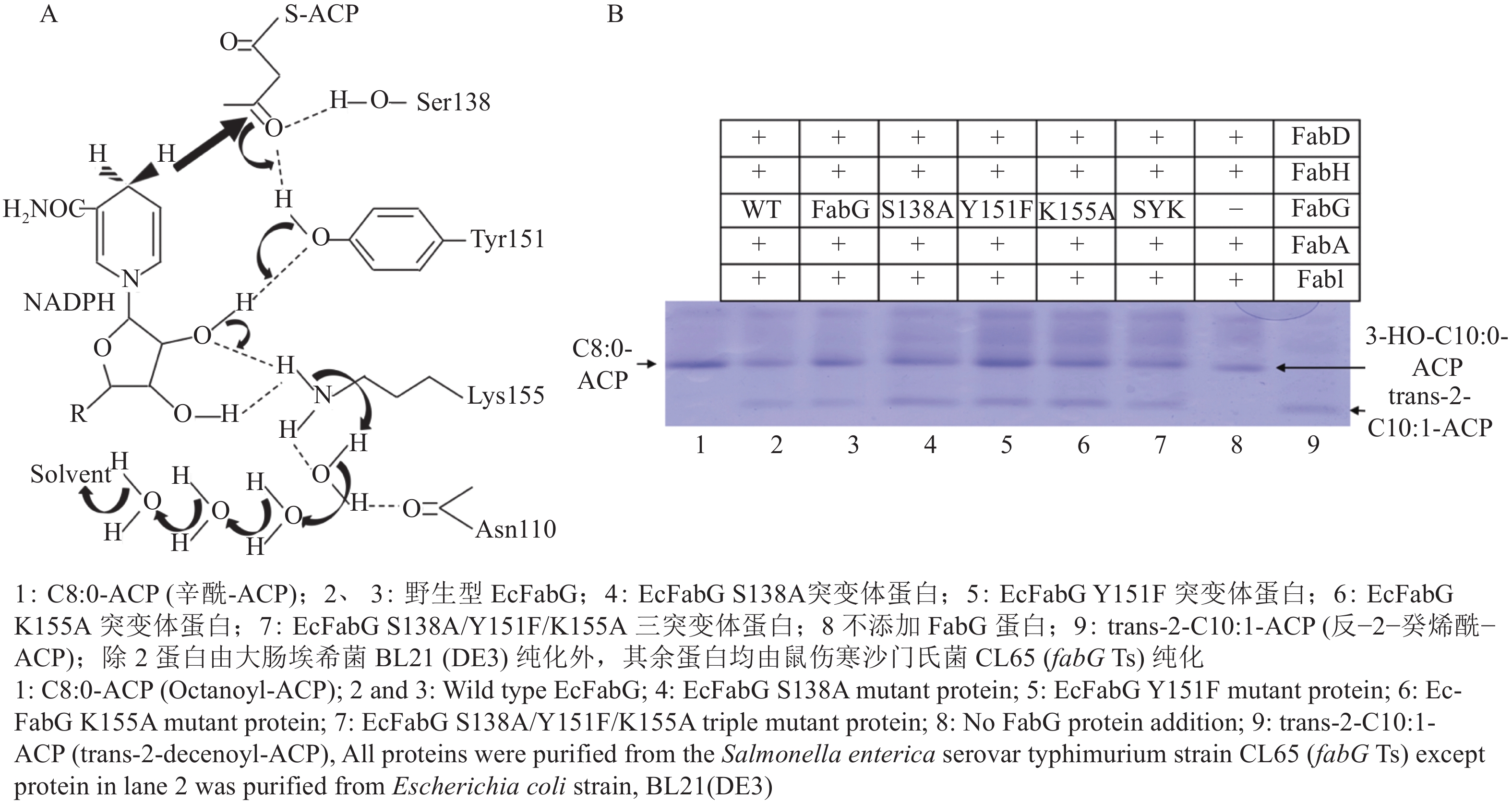
Price等[54]进一步解析了FabG的活性位点,Tyr229在空间上位于α6/α7螺旋相对的亲水口袋中,参与了氢键网络的形成,Arg129和Arg172在脂酰ACP和FabG的相互作用上起着重要作用。Price等[54]证明 Ser138、Tyr151和Lys155的突变体蛋白与NADPH的结合能力减弱、对乙酰乙酰辅酶A的催化活性急剧降低。保守基序Ser 138,Tyr151和Lys155是必需氨基酸,均集中在α5/β5催化口袋附近,在NADPH结合和电子传递中起着重要的作用。NADPH的结合会增强FabG对脂酰ACP的亲和力,降低最大结合浓度,引起蛋白构象的变化,导致Ser138、Tyr151和Lys155的重排。NADPH中核糖结构上的羟基、Ser-Tyr-Lys基序和四分子水形成质子传递通路。质子由NADPH提供,Tyr151羟基介导,最终完成3−酮脂酰ACP还原(图4A)[58]。
Ser138、Tyr151和Lys155突变体蛋白虽然对乙酰乙酰辅酶A的催化活性急剧降低,但FabG的天然底物是3−酮脂酰ACP而非脂酰辅酶A[59]。对茄科雷尔氏菌FabG1的研究表明突变Ser-Tyr-Lys任意残基并不会丧失OAR的功能,在42 ℃依然能够恢复大肠埃希菌CL104生长[42]。Hu等[60]对大肠埃希菌FabG催化活性位点展开研究,发现Ser138Ala、Tyr151Phe和Lys155Ala突变体蛋白在体外和体内都保有足够的OAR催化活性(图4B)。突变体蛋白不仅能互补大肠埃希菌CL104菌株,而且能互补鼠伤寒沙门氏菌温度敏感突变菌株CL65。在CL65中纯化表达突变体蛋白,突变体蛋白在体外能够在起始反应和延伸反应中完成脂肪酸合成。进一步以3−酮脂酰ACP为底物测定突变体蛋白活性,发现相对于野生型FabG,Ser138Ala、Tyr151Phe和Lys155Ala分别有82%、59%和50%的活力。这表明Ser138、Tyr151和Lys155并不是EcFabG的催化活性中心,更可能是作为维持催化中心结构的氨基酸存在。以上研究结果表明,OAR可能存在不同于典型SDR超家族蛋白的电子传递机制和保守催化活性中心。
2.2 细菌3−酮脂酰ACP还原酶的抑制剂研究
相比于细菌脂肪酸合成系统的其他成员,OAR在不同细菌间较为保守,是非常优良的广谱抗生素开发靶位点[61]。有关筛选细菌OAR抑制剂的工作也取得积极进展[62-63]。
植物多酚类物质,尤其是黄酮类化合物对OAR具有抑制效果,如绿茶中的焙儿茶素(Epigallocatechin gallate,EGCG)及儿茶素是优良的抑菌剂。EGCG及儿茶素以FAS II系统作为抑制靶点,有效地抑制了脂肪酸延伸过程中FabG和FabI所催化的还原反应[64]。单宁酸及枫叶提取物革兰阳性菌的抗菌活性也来自对OAR的抑制[65]。有研究表明,类黄酮和大环内酰亚胺对FabG有抑制作用,且为非特异性抑制,所以不能应用于临床[66];鲎素能够抑制4种革兰阴性细菌的OAR,但存在细胞毒性和溶血作用[67]。反式肉桂酸及其衍生物也有很显著的OAR抑制能力,这些化合物不仅能阻遏NADPH结合,还能够与多个重要的氨基酸活性残基结合,阻止底物的靠近[68]。
Cukier等[46]筛选到16个铜绿假单胞菌FabG蛋白抑制物,其IC50仅为nmol级,是极具潜力的铜绿假单胞菌抗菌药物,对这些抑制物的结合位点进行分析,发现抑制剂均结合在FabG亚基相邻界面的变构位点;此外他们还发现抑制剂在FabG的结合主要依赖于疏水相互作用,但也有抑制剂依赖于特定的氢键与FabG结合。抑制剂一旦与FabG结合,蛋白–抑制剂复合物的构象发生改变,无法与NADPH结合[24]。然而这些化合物对其他OAR的抑制效果并不好。
Vella等[56]筛选出一系列IC50在μmol级的抑制物,并且证明了这些抑制物与铜绿假单胞菌筛选到的抑制物一样,结合在亚基界面的变构位点上,尤其是α4/α5螺旋;通过对变构位点的研究证明Asn86羰基、侧链基团Tyr151和Lys155参与结合NAPDH的氢键形成,而多肽链骨架上的Gly182和Ile184也可能参与;抑制剂的结合位点主要由疏水残基组成,通过一个额外的极性相互作用作用于亚基表面,抑制物主要阻遏NADPH和OAR的相互作用。
Varakala等[67]通过高通量从细菌中筛选出43个可以抑制多个FabG的合成物,其中14个为吲哚类,29个为酰胺类;对药物动力学的研究表明,其中7个可以在μmol级抑制多个FabG的活性;5–溴–2–(噻吩–2–甲酰胺)苯甲酸和6–溴–2–(二甲氨基)甲基)–5–羟基–1–苯基–1H–吲哚–3–甲酸乙酯能够抑制6个FabG,且IC50为7~70 μmol/L,2–(2,6–二氯苄基)硫代)吡唑[1,5–a][1,3,5]三嗪–4(3H)–酮能够抑制4个FabG。
3. 其他类型的3–酮脂酰ACP还原酶
许多细菌基因组中,除了fab基因簇的fabG基因外,还有多个基因被标注为OAR编码基因。这些基因大体可分为2类:一类编码的蛋白在体外和体内均具有OAR活性,但它们不参与脂肪酸合成,而是专一地参与一些次生代谢物的合成,执行特殊的生物学功能;另一类编码的蛋白没有OAR活性,具体的生物学功能有待深入研究。
3.1 分枝杆菌的MabA参与分枝酸的合成
分枝酸是分枝杆菌属细菌细胞壁的特殊成分,是一类含60~90个碳原子的长链3–羟基脂肪酸。研究证明分枝杆菌的膜磷脂脂肪酸由I型脂肪酸合成系统(FAS I)生成,而分枝酸的合成则由FAS II催化。分枝杆菌首先通过FAS I以乙酰–CoA为底物合成16~26碳的脂肪酸链,接下来利用FAS II将碳链进一步延伸到48~60个碳,进而形成分枝酸。分枝杆菌的FAS II 相关基因包括fabD-acpM-kasA-kasB-accA基因簇和mabA-inhA基因簇。
Banerjee等[69]克隆了结核分枝杆菌的mabA(Rv1483,MtFabG1),并纯化得到MabA蛋白,MabA蛋白能够消耗NADPH,并还原乙酰乙酰辅酶A。这表明MabA蛋白具有OAR活性。Marrakchi等[70]证实MabA参与结核分枝杆菌的分枝酸合成,对短链的3–酮脂酰脂肪酸的亲和力很低,而对长链(C8~C16)的3–酮脂酰脂肪酸的亲和力较高。Parish等[71]证实inhA与mabA共转录;且只有在外源携带有编码mabA-inhA基因簇的质粒存在时才能将染色体上的mabA敲除,这表明mabA是必需基因。此外Parish等[71]也发现大肠埃希菌fabG并不能替换结核分枝杆菌的mabA,而耻垢分枝杆菌Mycobacterium smegmatis的mabA可以替换结核分枝杆菌mabA,并且替换菌株的分枝酸和细胞通透性没有改变。这表明分枝杆菌FAS II系统有独特的OAR,参与分枝酸的合成。
3.2 苜蓿中华根瘤菌NodG参与结瘤因子合成
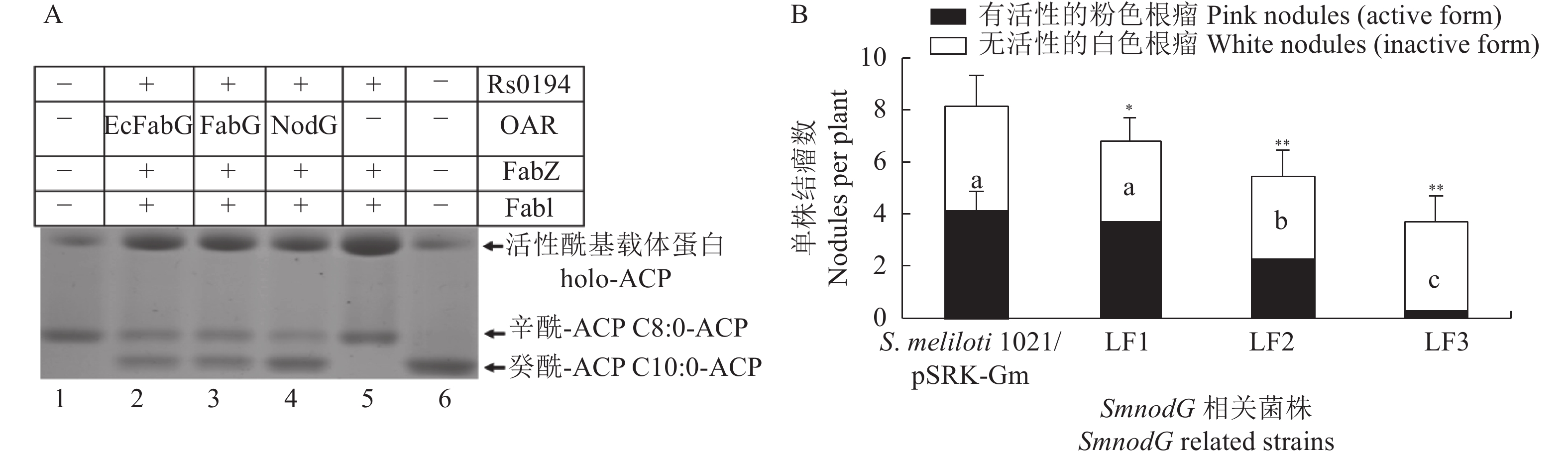
除染色体外,苜蓿中华根瘤菌还有1个巨质粒SymA,与共生结瘤相关的nod基因簇位于该巨质粒上。nod基因簇中的nodG被标注为OAR,其编码的蛋白与大肠埃希菌FabG的序列一致性达到53%。nodG不是苜蓿中华根瘤生长的必需基因,敲除后并不影响菌体脂肪酸生成。但是体外和体内试验显示NodG具有OAR活性(图5A)。同时证明,在过表达nodG的情况下可以敲除fabG,且突变菌株的脂肪酸组成与野生菌株一致。这表明nodG在苜蓿中华根瘤菌中行使与fabG相似的功能。然而遗传学的研究发现nodG缺失会导致苜蓿结瘤效率降低,并证明其专一性地参与结瘤因子的合成(图5B)[43]。
![图 5 苜蓿中华根瘤菌NodG具有OAR活性并参与苜蓿结瘤]() 图 5 苜蓿中华根瘤菌NodG具有OAR活性并参与苜蓿结瘤A:苜蓿根瘤菌FabG与NodG具有OAR活性;1:C8:0-ACP,2:EcFabG,3:苜蓿根瘤菌FabG,4:苜蓿根瘤菌NodG,5:不添加FabG蛋白,6:C8:0-ACP。B:NodG相关菌株结瘤效率;S. meliloti 1021/pSRK-Gm:野生型菌株携带空质粒,LF1:苜蓿根瘤菌nodG突变菌株,LF2:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌nodG质粒,LF3:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌fabG质粒, “*”“**”分别表示0.05和0.01水平差异显著[43],每个试验有8个重复Figure 5. The Sinorhizobium meliloti NodG maintains OAR activity and involves in alfalfa nodulationA: Sinorhizobium meliloti FabG and NodG maintain the OAR activity; 1: C8:0-ACP, 2: Product of EcFabG, 3: Product of SmFabG, 4: Product of SmNodG, 5: No FabG addition, 6: C8:0-ACP. B: Nodulation efficiency of Sinorhizobium meliloti mutant strains; S. meliloti 1021/pSRK-Gm: Wild type strain carrying plasmid pSRK-Gm, LF1: Sinorhizobium meliloti nodG mutant strain, LF2: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmnodG ecoding plasmid, LF3: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmfabG ecoding plasmid, “*” and “**”indicate significant differences at levels of 0.05 and 0.01, respectively, Every experiment has eight replicates
图 5 苜蓿中华根瘤菌NodG具有OAR活性并参与苜蓿结瘤A:苜蓿根瘤菌FabG与NodG具有OAR活性;1:C8:0-ACP,2:EcFabG,3:苜蓿根瘤菌FabG,4:苜蓿根瘤菌NodG,5:不添加FabG蛋白,6:C8:0-ACP。B:NodG相关菌株结瘤效率;S. meliloti 1021/pSRK-Gm:野生型菌株携带空质粒,LF1:苜蓿根瘤菌nodG突变菌株,LF2:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌nodG质粒,LF3:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌fabG质粒, “*”“**”分别表示0.05和0.01水平差异显著[43],每个试验有8个重复Figure 5. The Sinorhizobium meliloti NodG maintains OAR activity and involves in alfalfa nodulationA: Sinorhizobium meliloti FabG and NodG maintain the OAR activity; 1: C8:0-ACP, 2: Product of EcFabG, 3: Product of SmFabG, 4: Product of SmNodG, 5: No FabG addition, 6: C8:0-ACP. B: Nodulation efficiency of Sinorhizobium meliloti mutant strains; S. meliloti 1021/pSRK-Gm: Wild type strain carrying plasmid pSRK-Gm, LF1: Sinorhizobium meliloti nodG mutant strain, LF2: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmnodG ecoding plasmid, LF3: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmfabG ecoding plasmid, “*” and “**”indicate significant differences at levels of 0.05 and 0.01, respectively, Every experiment has eight replicates3.3 野油菜黄单胞菌FabG2是一种新型OAR
可扩散信号分子DSF介导黄单胞菌的群体感应调控,目前在黄单胞菌中已经鉴定到4种DSF类信号,它们的化学本质是主碳链含12个碳原子的顺–2–不饱和烯酸[72-73]。Yu等[13]研究表明野油菜黄单胞菌的3–酮脂酰ACP合成酶III能以支链CoA为底物合成支链脂肪酸,同时决定了野油菜黄单胞菌产生多种类型的DSF类信号。
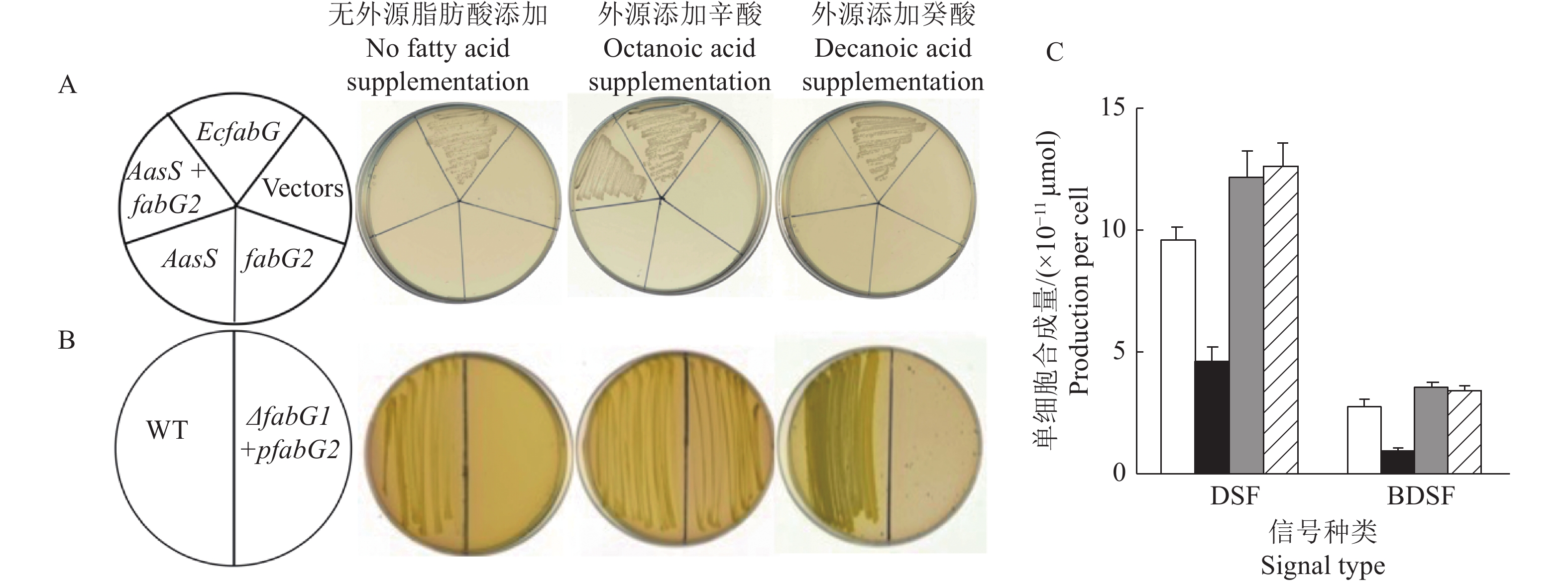
在野油菜黄单胞菌中,与fabG1不同,fabG2是1个独立基因,其编码的蛋白与大肠埃希菌FabG的序列一致性仅为32.4%。Hu等[44]的研究发现FabG2对短链3–酮脂酰ACP的催化活性低,单独fabG2不能恢复大肠埃希菌fabG温度敏感突变菌CL104的生长,也不参与野油菜黄单胞菌脂肪酸的从头合成。然而在与哈氏弧菌Vibrio harveyi脂酰ACP合成酶基因(aasS)共转录,并外源添加辛酸后,fabG2可以使CL104在非许可温度生长,表明FabG2对中长链的3–酮脂酰ACP具有较强的催化活性(图6A)。另外研究发现,在添加有辛酸时,过表达fabG2能够获得fabG1敲除突变菌株(图6B)。更为重要的是敲除XccfabG2导致野油菜黄单胞菌的DSF类信号合成量显著降低(图6C)。这表明FabG2是1个专一性参与DSF类信号合成调控的新型OAR。
![图 6 XccfabG2互补OAR突变菌株及参与Xcc DSF类信号分子合成]() 图 6 XccfabG2互补OAR突变菌株及参与Xcc DSF类信号分子合成A:XccfabG2互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,Vector:空质粒载体,fabG2:质粒携带有XccfabG2基因,AasS:质粒携带有哈氏弧菌aasS基因,AasS+ fabG:质粒携带有哈氏弧菌aasS和XccfabG2基因。B:XccfabG2互补XccfabG1突变菌株;WT:Xcc野生型,∆fabG1+pfabG2:XccfabG1突变菌株携带有XccfabG2表达质粒。C: XccfabG2突变菌株的DSF类信号分子产量;DSF:11−甲基−顺−2−月桂酰烯酸,BDSF:顺−2−月桂酰烯酸,空柱:Xcc野生型,黑柱:XccfabG2突变菌株,灰柱:XccfabG2突变菌株携带有XccfabG2表达质粒,条状柱:XccfabG2突变菌株携带有XccfabG1表达质粒Figure 6. XccfabG2 complements OAR mutant strain and invovles in DSF signal synthesis in XccA: XccfabG2 complement E. coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, Vector: CL104 carrying empty plasmid, fabG2: CL104 carrying plasmid expressing XccfabG2, AasS: CL104 carrying plasmid expressing Vibrio harzii aasS, AasS+ fabG2: CL104 carrying plasmid expressing Vibrio harzii aasS and XccfabG2. B: XccfabG2 complement XccfabG1 mutant strain; WT: Wild type X. campestris pv. campestris strain, ∆fabG1+pfabG2: XccfabG1 mutant strain carrying XccfabG2 expressing plasmid. C: The DSFs production of XccfabG2 mutant strains; DSF: cis-11-methyl-2-dodecenoic acid, BDSF: cis-2-dodecenoic acid, White column: Wild-type X. campestris pv. campestris strain, Black column: XccfabG2 mutant strain, Gray column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG2, Stippled column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG1
图 6 XccfabG2互补OAR突变菌株及参与Xcc DSF类信号分子合成A:XccfabG2互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,Vector:空质粒载体,fabG2:质粒携带有XccfabG2基因,AasS:质粒携带有哈氏弧菌aasS基因,AasS+ fabG:质粒携带有哈氏弧菌aasS和XccfabG2基因。B:XccfabG2互补XccfabG1突变菌株;WT:Xcc野生型,∆fabG1+pfabG2:XccfabG1突变菌株携带有XccfabG2表达质粒。C: XccfabG2突变菌株的DSF类信号分子产量;DSF:11−甲基−顺−2−月桂酰烯酸,BDSF:顺−2−月桂酰烯酸,空柱:Xcc野生型,黑柱:XccfabG2突变菌株,灰柱:XccfabG2突变菌株携带有XccfabG2表达质粒,条状柱:XccfabG2突变菌株携带有XccfabG1表达质粒Figure 6. XccfabG2 complements OAR mutant strain and invovles in DSF signal synthesis in XccA: XccfabG2 complement E. coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, Vector: CL104 carrying empty plasmid, fabG2: CL104 carrying plasmid expressing XccfabG2, AasS: CL104 carrying plasmid expressing Vibrio harzii aasS, AasS+ fabG2: CL104 carrying plasmid expressing Vibrio harzii aasS and XccfabG2. B: XccfabG2 complement XccfabG1 mutant strain; WT: Wild type X. campestris pv. campestris strain, ∆fabG1+pfabG2: XccfabG1 mutant strain carrying XccfabG2 expressing plasmid. C: The DSFs production of XccfabG2 mutant strains; DSF: cis-11-methyl-2-dodecenoic acid, BDSF: cis-2-dodecenoic acid, White column: Wild-type X. campestris pv. campestris strain, Black column: XccfabG2 mutant strain, Gray column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG2, Stippled column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG13.4 野油菜黄单胞菌FabG3参与菌黄素合成
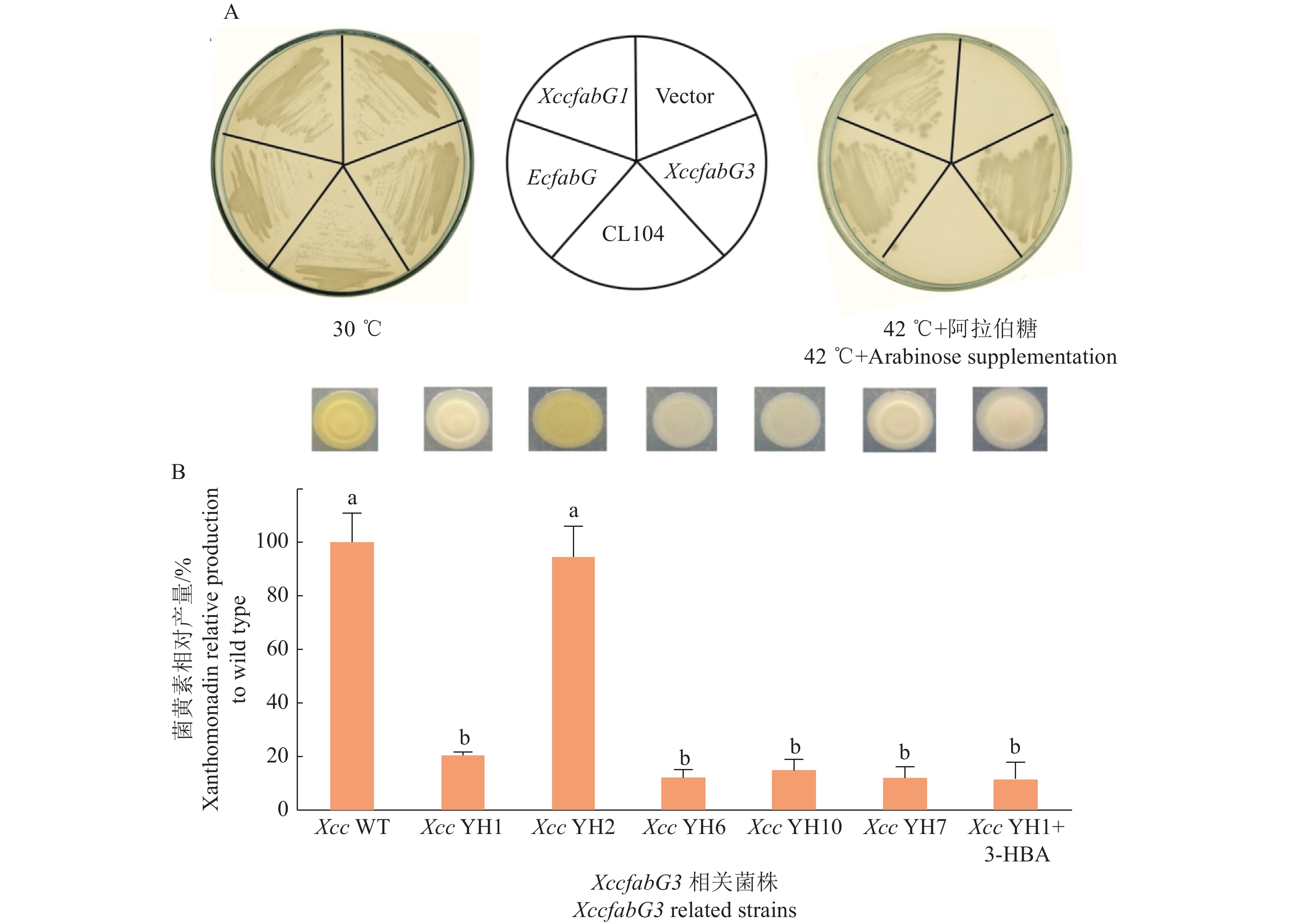
在野油菜黄单胞菌野油菜致病变种中,已经鉴定了3个OAR的生物学功能。FabG1参与脂肪酸合成,FabG2参与DSF类信号分子合成调控,而fabG3位于菌黄素合成基因簇中。Yu等[74]的研究结果表明,FabG3在体外和体内均能行使OAR的功能(图7A),但fabG3的缺失并不会导致Xcc的生长或脂肪酸组成发生改变;然而突变菌株菌黄素合成受阻,同时证明这一性状不能由其他OAR编码基因互补恢复,只能由fabG3互补恢复(图7B)。这些研究结果表明,FabG3是一种专一性参与菌黄素合成的OAR。
![图 7 XccfabG3互补CL104并参与菌黄素合成]() 图 7 XccfabG3互补CL104并参与菌黄素合成A:XccfabG3互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,fabG1:质粒携带有XccfabG1基因,Vector:空质粒载体,fabG3:质粒携带有XccfabG3基因,CL104:无外源质粒。B: XccfabG3突变菌株的菌黄素产量;Xcc WT:Xcc野生型,Xcc YH1:XccfabG3突变菌株,Xcc YH2:XccfabG3突变菌株携带有XccfabG3表达质粒,Xcc YH6:XccfabG3突变菌株携带有XccfabG1表达质粒,Xcc YH10:XccfabG3突变菌株携带有XccfabG2表达质粒,Xcc YH7:XccfabG3突变菌株携带有EcfabG表达质粒,Xcc YH1+3-HBA:XccfabG3突变菌株添加3−羟基丁酸Figure 7. XccfabG3 complements CL104 and invovles in xanthomonadin synthesis in XccA: XccfabG3 complement Escherichia coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, fabG1: CL104 carrying plasmid expressing XccfabG1, Vector: CL104 carrying empty plasmid, fabG3: CL104 carrying plasmid expressing XccfabG3, CL104: CL104 without plasmid. B: Xanthomonadin production in XccfabG3 mutant; Xcc WT: Wild-type X. campestris pv. campestris strain, Xcc YH1: XccfabG3 mutant, Xcc YH2: XccfabG3 mutant carrying XccfabG3 ecoding plasmid, Xcc YH6: XccfabG3 mutant carrying XccfabG1 ecoding plasmid, Xcc YH10: XccfabG3 mutant carrying XccfabG2 ecoding plasmid, Xcc YH7: XccfabG3 mutant carrying EcfabG ecoding plasmid, Xcc YH1+3-HBA: XccfabG3 mutant supplemented with 3-hydroxybutyric acid
图 7 XccfabG3互补CL104并参与菌黄素合成A:XccfabG3互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,fabG1:质粒携带有XccfabG1基因,Vector:空质粒载体,fabG3:质粒携带有XccfabG3基因,CL104:无外源质粒。B: XccfabG3突变菌株的菌黄素产量;Xcc WT:Xcc野生型,Xcc YH1:XccfabG3突变菌株,Xcc YH2:XccfabG3突变菌株携带有XccfabG3表达质粒,Xcc YH6:XccfabG3突变菌株携带有XccfabG1表达质粒,Xcc YH10:XccfabG3突变菌株携带有XccfabG2表达质粒,Xcc YH7:XccfabG3突变菌株携带有EcfabG表达质粒,Xcc YH1+3-HBA:XccfabG3突变菌株添加3−羟基丁酸Figure 7. XccfabG3 complements CL104 and invovles in xanthomonadin synthesis in XccA: XccfabG3 complement Escherichia coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, fabG1: CL104 carrying plasmid expressing XccfabG1, Vector: CL104 carrying empty plasmid, fabG3: CL104 carrying plasmid expressing XccfabG3, CL104: CL104 without plasmid. B: Xanthomonadin production in XccfabG3 mutant; Xcc WT: Wild-type X. campestris pv. campestris strain, Xcc YH1: XccfabG3 mutant, Xcc YH2: XccfabG3 mutant carrying XccfabG3 ecoding plasmid, Xcc YH6: XccfabG3 mutant carrying XccfabG1 ecoding plasmid, Xcc YH10: XccfabG3 mutant carrying XccfabG2 ecoding plasmid, Xcc YH7: XccfabG3 mutant carrying EcfabG ecoding plasmid, Xcc YH1+3-HBA: XccfabG3 mutant supplemented with 3-hydroxybutyric acid3.5 其他OAR编码基因功能
铜绿假单胞菌是一种重要的条件致病菌,脂肪酸合成系统在铜绿假单胞菌的多种致病过程中起重要作用。在PAO1的基因组中,有42个SDR超家族基因,12个标注为OAR家族基因。Guo等[45]对铜绿假单胞菌基因组中的候选基因进行了系统研究,结果发现除fabG外,只有PA4389和PA4786具有OAR的活性;PA4389和PA4786在体内和体外均能行使OAR功能。然而,PA4389和PA4786并不参与铜绿假单胞菌的脂肪酸合成,这2个基因突变后,铜绿假单胞菌的脂肪酸组成并没有显著改变,但信号分子2–庚基–3–羟基–4–喹诺酮类(PQS)合成量减少50%以上。这表明PA4389和PA4786虽然不直接参与铜绿假单胞菌的脂肪酸合成,却影响其信号分子的合成。
Guo等[45]研究表明其他的OAR家族候选基因虽然不具备OAR活性,但在其他代谢途径中发挥作用。PA3128与苜蓿中华根瘤菌参与TCA循环的关键基因SMc02486的氨基酸序列有60%的一致性。PA3128被敲除后,突变菌株的PQS合成量提高了20%,弹性蛋白酶水解酶LasB的活性降低20%。在基础培养基上,以L–丙氨酸或D–海藻糖为碳源时,PA3128的突变菌株生长缓慢,最终得到的生物量仅为野生型的20%~40%,而以D–天冬氨酸为唯一碳源时,PA3128的突变菌株完全不能生长。以上结果表明PA3128可能也参与铜绿假单胞菌的TCA循环。而PA0182和PA1470在铜绿假单胞菌利用L–丝氨酸、L–天冬氨酸、L–谷氨酸和L–组氨酸时发挥重要作用,任何单突变菌株均不能利用以上氨基酸。PA3387(RhlG)曾认为参与铜绿假单胞菌重要致病因子鼠李糖脂的合成,然而Zhu等[33]的研究表明无论在体外还是体内RhlG都不能催化3–酮基癸酰ACP还原为3–羟基癸酰ACP,并不参与鼠李糖脂的合成。
在天蓝色链霉菌Streptomyces coelicolor中,脂肪酸可以用于次级代谢产物十二烷基二苷酸的生物合成。天蓝色链霉菌基因组的3个OAR候选基因编码的蛋白均能够催化β–酮脂酰–N–乙酰半胱氨酸。其中SOC1815是脂肪酸合成的关键酶;而SOC1345能够区分β–酮脂酰–N–乙酰半胱氨酸–ACP和β–酮脂酰–ACP;SOC1846的生物学功能尚不清楚[75]。
4. 总结与展望
细菌脂肪酸合成机制高度保守,这不仅表现在不同细菌脂肪酸合成相关酶的蛋白序列高度相似,而且相关编码基因在染色体上的排列顺序也非常相似。但是研究发现不同细菌的脂肪酸合成机制存在差异,主要体现在:1)同种细菌中催化同一生化反应的酶有多种;2)不同细菌催化同一生化反应的酶的结构不同;3)不同细菌合成同样产物的方式和机制不同。
生物信息学分析显示细菌基因组中有许多注释为OAR的编码基因,这些基因编码的蛋白有些具有OAR活性,且能够遗传互补大肠埃希菌fabG温敏突变菌株,但是研究证明真正参与细菌脂肪酸合成的OAR由位于脂肪酸合成基因簇(fab)中的fabG编码。fabG是细菌的必需基因,突变该基因致死细菌。虽然有些OAR的编码基因,例如中华苜蓿根瘤菌nodG和野油菜黄单胞菌fabG2,在某些特殊条件下能够替代fabG,合成脂肪酸,维持菌体生长,但是NodG和FabG2不是FabG的同工酶,它们各自有自己的生物功能,催化不同物质的合成。因此,与参与细菌脂肪酸合成的其他酶不同,OAR只有FabG类同工酶,因此,FabG是一个理想的抗菌药物筛选靶标,可用于广谱抗菌药物的筛选。
除了参与细菌脂肪酸合成的FabG外,细菌中还有一些具有OAR活性的蛋白,目前研究证明这些OAR往往专一地参与脂肪酸相关物质的合成,如分枝酸、结瘤因子、菌黄素等。这表明这些酶对3–酮基脂酰基团具有还原能力。因此,对这些OAR功能的鉴定将有助于研究一些与脂肪酸相关物质合成,至少可以推测这些物质合成中可能有3–酮基脂酰基团的还原反应。
FabG作为一个潜在的优良广谱抗菌药物的开发靶点,虽然引起了科学家们的高度关注,并据此筛选或设计了一些药物,但目前鲜见临床应用或工业化生产的报道。Ser-Tyr-Lys基序是SDR超家族蛋白的结构特征之一,也是该类蛋白的催化活性中心,然而大肠埃希菌FabG催化机制研究证明,Ser-Tyr-Lys基序在FabG催化3–酮基脂酰ACP还原中并不起关键作用,这表明FabG可能采用有别于一般SDR家族蛋白的催化机制。这也要求对FabG更深入地研究,进一步解析FabG的结构特点和催化特性,为以FabG为靶点开展药物设计奠定基础。
-
图 5 苜蓿中华根瘤菌NodG具有OAR活性并参与苜蓿结瘤
A:苜蓿根瘤菌FabG与NodG具有OAR活性;1:C8:0-ACP,2:EcFabG,3:苜蓿根瘤菌FabG,4:苜蓿根瘤菌NodG,5:不添加FabG蛋白,6:C8:0-ACP。B:NodG相关菌株结瘤效率;S. meliloti 1021/pSRK-Gm:野生型菌株携带空质粒,LF1:苜蓿根瘤菌nodG突变菌株,LF2:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌nodG质粒,LF3:苜蓿根瘤菌nodG/fabG双突变菌株携带苜蓿根瘤菌fabG质粒, “*”“**”分别表示0.05和0.01水平差异显著[43],每个试验有8个重复
Figure 5. The Sinorhizobium meliloti NodG maintains OAR activity and involves in alfalfa nodulation
A: Sinorhizobium meliloti FabG and NodG maintain the OAR activity; 1: C8:0-ACP, 2: Product of EcFabG, 3: Product of SmFabG, 4: Product of SmNodG, 5: No FabG addition, 6: C8:0-ACP. B: Nodulation efficiency of Sinorhizobium meliloti mutant strains; S. meliloti 1021/pSRK-Gm: Wild type strain carrying plasmid pSRK-Gm, LF1: Sinorhizobium meliloti nodG mutant strain, LF2: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmnodG ecoding plasmid, LF3: Sinorhizobium meliloti nodG/fabG double mutant strain carrying SmfabG ecoding plasmid, “*” and “**”indicate significant differences at levels of 0.05 and 0.01, respectively, Every experiment has eight replicates
图 6 XccfabG2互补OAR突变菌株及参与Xcc DSF类信号分子合成
A:XccfabG2互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,Vector:空质粒载体,fabG2:质粒携带有XccfabG2基因,AasS:质粒携带有哈氏弧菌aasS基因,AasS+ fabG:质粒携带有哈氏弧菌aasS和XccfabG2基因。B:XccfabG2互补XccfabG1突变菌株;WT:Xcc野生型,∆fabG1+pfabG2:XccfabG1突变菌株携带有XccfabG2表达质粒。C: XccfabG2突变菌株的DSF类信号分子产量;DSF:11−甲基−顺−2−月桂酰烯酸,BDSF:顺−2−月桂酰烯酸,空柱:Xcc野生型,黑柱:XccfabG2突变菌株,灰柱:XccfabG2突变菌株携带有XccfabG2表达质粒,条状柱:XccfabG2突变菌株携带有XccfabG1表达质粒
Figure 6. XccfabG2 complements OAR mutant strain and invovles in DSF signal synthesis in Xcc
A: XccfabG2 complement E. coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, Vector: CL104 carrying empty plasmid, fabG2: CL104 carrying plasmid expressing XccfabG2, AasS: CL104 carrying plasmid expressing Vibrio harzii aasS, AasS+ fabG2: CL104 carrying plasmid expressing Vibrio harzii aasS and XccfabG2. B: XccfabG2 complement XccfabG1 mutant strain; WT: Wild type X. campestris pv. campestris strain, ∆fabG1+pfabG2: XccfabG1 mutant strain carrying XccfabG2 expressing plasmid. C: The DSFs production of XccfabG2 mutant strains; DSF: cis-11-methyl-2-dodecenoic acid, BDSF: cis-2-dodecenoic acid, White column: Wild-type X. campestris pv. campestris strain, Black column: XccfabG2 mutant strain, Gray column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG2, Stippled column: XccfabG2 mutant strain carrying a plasmid encoding XccfabG1
图 7 XccfabG3互补CL104并参与菌黄素合成
A:XccfabG3互补大肠埃希菌fabG温度敏感突变菌株CL104;EcfabG:质粒携带有大肠埃希菌fabG基因,fabG1:质粒携带有XccfabG1基因,Vector:空质粒载体,fabG3:质粒携带有XccfabG3基因,CL104:无外源质粒。B: XccfabG3突变菌株的菌黄素产量;Xcc WT:Xcc野生型,Xcc YH1:XccfabG3突变菌株,Xcc YH2:XccfabG3突变菌株携带有XccfabG3表达质粒,Xcc YH6:XccfabG3突变菌株携带有XccfabG1表达质粒,Xcc YH10:XccfabG3突变菌株携带有XccfabG2表达质粒,Xcc YH7:XccfabG3突变菌株携带有EcfabG表达质粒,Xcc YH1+3-HBA:XccfabG3突变菌株添加3−羟基丁酸
Figure 7. XccfabG3 complements CL104 and invovles in xanthomonadin synthesis in Xcc
A: XccfabG3 complement Escherichia coli fabG temperature sensitive strain CL104; EcfabG: CL104 carrying plasmid expressing EcfabG, fabG1: CL104 carrying plasmid expressing XccfabG1, Vector: CL104 carrying empty plasmid, fabG3: CL104 carrying plasmid expressing XccfabG3, CL104: CL104 without plasmid. B: Xanthomonadin production in XccfabG3 mutant; Xcc WT: Wild-type X. campestris pv. campestris strain, Xcc YH1: XccfabG3 mutant, Xcc YH2: XccfabG3 mutant carrying XccfabG3 ecoding plasmid, Xcc YH6: XccfabG3 mutant carrying XccfabG1 ecoding plasmid, Xcc YH10: XccfabG3 mutant carrying XccfabG2 ecoding plasmid, Xcc YH7: XccfabG3 mutant carrying EcfabG ecoding plasmid, Xcc YH1+3-HBA: XccfabG3 mutant supplemented with 3-hydroxybutyric acid
-
[1] PARSONS J B, ROCK C O. Bacterial lipids: Metabolism and membrane homeostasis[J]. Progress in Lipid Research, 2013, 52(3): 249-276. doi: 10.1016/j.plipres.2013.02.002
[2] KANEDA T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance[J]. Microbiological Reviews, 1991, 55(2): 288-302. doi: 10.1128/mr.55.2.288-302.1991
[3] CRONAN J E, THOMAS J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways[J]. Methods in Enzymology, 2009, 459: 395-433.
[4] SANJURJO P, RUIZ J I, MONTEJO M. Inborn errors of metabolism with a protein-restricted diet: Effect on polyunsaturated fatty acids[J]. Journal of Inherited Metabolic Disease, 1997, 20(6): 783-789. doi: 10.1023/A:1005367701176
[5] ROCK C O, CRONAN J E. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis[J]. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism, 1996, 1302(1): 1-16. doi: 10.1016/0005-2760(96)00056-2
[6] GOVERS-RIEMSLAG J W P, JANSSEN M P, ZWAAL R F A, et al. Effect of membrane fluidity and fatty acid composition on the prothrombin-converting activity of phospholipid vesicles[J]. Biochemistry, 1992, 31(41): 10000-10008.
[7] ZHANG Y M, ROCK C O. Membrane lipid homeostasis in bacteria[J]. Nature Reviews Microbiology, 2008, 6(3): 222-233. doi: 10.1038/nrmicro1839
[8] YUAN Y Q, LEEDS J A, MEREDITH T C. Pseudomonas aeruginosa directly shunts β-oxidation degradation intermediates into de novo fatty acid biosynthesis[J]. Journal of Bacteriology, 2012, 194(19): 5185-5196. doi: 10.1128/JB.00860-12
[9] ZHANG L, VERES-SCHALNAT T A, SOMOGYI A, et al. Fatty acid cosubstrates provide β-oxidation precursors for rhamnolipid biosynthesis in Pseudomonas aeruginosa, as evidenced by isotope tracing and gene expression assays[J]. Applied and Environmental Microbiology, 2012, 78(24): 8611-8622. doi: 10.1128/AEM.02111-12
[10] CHRISTENSEN Q H, CRONAN J E. Lipoic acid synthesis: A new family of octanoyltransferases generally annotated as lipoate protein ligases[J]. Biochemistry, 2010, 49(46): 10024-10036. doi: 10.1021/bi101215f
[11] LIN S, HANSON R E, CRONAN J E. Biotin synthesis begins by hijacking the fatty acid synthetic pathway[J]. Nature Chemical Biology, 2010, 6(9): 682-688. doi: 10.1038/nchembio.420
[12] WANG Q, LIU C S, XIAN M, et al. Biosynthetic pathway for poly(3-hydroxypropionate) in recombinant Escherichia coli[J]. Journal of Microbiology, 2012, 50(4): 693-697. doi: 10.1007/s12275-012-2234-y
[13] YU Y, HU Z, DONG H, et al. Xanthomonas campestris FabH is required for branched-chain fatty acid and DSF-family quorum sensing signal biosynthesis[J]. Scientific Reports, 2016, 6: 32811. doi: 10.1038/srep32811
[14] HAN J, LU Q H, ZHOU L G, et al. Identification of the polyhydroxyalkanoate (PHA)-specific acetoacetyl coenzyme A reductase among multiple FabG paralogs in Haloarcula hispanica and reconstruction of the PHA biosynthetic pathway in Haloferax volcanii[J]. Applied and Environmental Microbiology, 2009, 75(19): 6168-6175. doi: 10.1128/AEM.00938-09
[15] CRONAN J E JR. The structure of mammalian fatty acid synthase turned back to front[J]. Chemistry & Biology, 2004, 11(12): 1601-1602.
[16] AMSTUTZ C L, FRISTEDT R, SCHULTINK A, et al. An atypical short-chain dehydrogenase-reductase functions in the relaxation of photoprotective qH in Arabidopsis[J]. Nature Plants, 2020, 6(2): 154-166. doi: 10.1038/s41477-020-0591-9
[17] KOO A J, FULDA M, BROWSE J, et al. Identification of a plastid acyl‐acyl carrier protein synthetase in Arabidopsis and its role in the activation and elongation of exogenous fatty acids[J]. The Plant Journal, 2005, 44(4): 620-632. doi: 10.1111/j.1365-313X.2005.02553.x
[18] LEE S, JUNG Y, LEE S, et al. Correlations between FAS elongation cycle genes expression and fatty acid production for improvement of long-chain fatty acids in Escherichia coli[J]. Applied Biochemistry and Biotechnology, 2013, 169(5): 1606-1619. doi: 10.1007/s12010-012-0088-8
[19] CRONAN J E JR. Phospholipid modifications in bacteria[J]. Current Opinion in Microbiology, 2002, 5(2): 202-205. doi: 10.1016/S1369-5274(02)00297-7
[20] MA J C, WU Y Q, CAO D, et al. Only acyl carrier protein 1 (AcpP1) functions in Pseudomonas aeruginosa fatty acid synthesis[J]. Frontiers in Microbiology, 2017, 8: 2186.
[21] ROCK C O, JACKOWSKI S. Forty years of bacterial fatty acid synthesis[J]. Biochemical and Biophysical Research Communications, 2002, 292(5): 1155-1166. doi: 10.1006/bbrc.2001.2022
[22] KONDAKOVA T, KUMAR S, CRONAN J E. A novel synthesis of trans-unsaturated fatty acids by the Gram-positive commensal bacterium Enterococcus faecalis FA2-2[J]. Chemistry and Physics of Lipids, 2019, 222: 23-35. doi: 10.1016/j.chemphyslip.2019.04.010
[23] HEATH R J, ROCK C O. Fatty acid biosynthesis as a target for novel antibacterials[J]. Current Opinion in Investigational Drugs, 2004, 5(2): 146-153.
[24] PARSONS J B, ROCK C O. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery?[J]. Current Opinion in Microbiology, 2011, 14(5): 544-549. doi: 10.1016/j.mib.2011.07.029
[25] MOLNOS J, GARDINER R, DALE G E, et al. A continuous coupled enzyme assay for bacterial malonyl-CoA: Acyl carrier protein transacylase (FabD)[J]. Analytical Biochemistry, 2003, 319(1): 171-176. doi: 10.1016/S0003-2697(03)00327-0
[26] SIX D A, YUAN Y Q, LEEDS J A, et al. Deletion of the β-acetoacetyl synthase FabY in Pseudomonas aeruginosa induces hypoacylation of lipopolysaccharide and increases antimicrobial susceptibility[J]. Antimicrobial Agents and Chemotherapy, 2014, 58(1): 153-161. doi: 10.1128/AAC.01804-13
[27] 余永红, 段园园, 董会娟, 等. 茄科雷尔氏菌脂酰−CoA合成酶的功能鉴定[J]. 微生物学通报, 2017, 44(2): 366-374. doi: 10.13344/j.microbiol.china.160688 [28] BI H, ZHU L, JIA J, et al. Unsaturated fatty acid synthesis in the gastric pathogen Helicobacter pylori proceeds via a backtracking mechanism[J]. Cell Chemical Biology, 2016, 23(12): 1480-1489. doi: 10.1016/j.chembiol.2016.10.007
[29] 雷鸣, 马金成, 王海洪. 流产布氏杆菌烯脂酰ACP还原酶的鉴定[J]. 生物化学与生物物理进展, 2012, 39(5): 464-471. [30] 余永红, 马建荣, 王海洪. 野油菜黄单胞菌中烯脂酰ACP还原酶的功能鉴定[J]. 生物化学与生物物理进展, 2016, 43(5): 514-522. doi: 10.16476/j.pibb.2015.0343 [31] DONG H J, MA J C, CHEN Q Y, et al. A cryptic long-chain 3-ketoacyl-ACP synthase in the Pseudomonas putida F1 unsaturated fatty acid synthesis pathway[J]. The Journal of Biological Chemistry, 2021, 297(2): 100920.
[32] HEATH R J, ROCK C O. Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis[J]. The Journal of Biological Chemistry, 1996, 271(44): 27795-27801. doi: 10.1074/jbc.271.44.27795
[33] ZHU L, CHENG J L, LUO B, et al. Functions of the Clostridium acetobutylicium FabF and FabZ proteins in unsaturated fatty acid biosynthesis[J]. BMC Microbiology, 2009, 9: 119.
[34] CAMPBELL J W, CRONAN J E JR. Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery[J]. Annual Review of Microbiology, 2001, 55: 305-332. doi: 10.1146/annurev.micro.55.1.305
[35] CHENG J L, MA J C, LIN J S, et al. Only one of the five Ralstonia solanacearum long-chain 3-ketoacyl-acyl carrier protein synthase homologues functions in fatty acid synthesis[J]. Applied and Environmental Microbiology, 2012, 78(5): 1563-1573. doi: 10.1128/AEM.07335-11
[36] GOLDBECK O, ECK A W, SEIBOLD G M. Real time monitoring of NADPH concentrations in Corynebacterium glutamicum and Escherichia coli via the genetically encoded sensor mBFP[J]. Frontiers in Microbiology, 2018, 9: 2564.
[37] KALLBERG Y, OPPERMANN U, PERSSON B. Classification of the short-chain dehydrogenase/reductase superfamily using hidden Markov models[J]. The FEBS Journal, 2010, 277(10): 2375-2386. doi: 10.1111/j.1742-4658.2010.07656.x
[38] ZHANG Y, CRONAN J. Transcriptional analysis of essential genes of the Escherichia coli fatty acid biosynthesis gene cluster by functional replacement with the analogous Salmonella typhimurium gene cluster[J]. Journal of Bacteriology, 1998, 180: 3295-3303.
[39] PODKOVYROV S M, LARSON T J. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli[J]. Nucleic Acids Research, 1996, 24(9): 1747-1752. doi: 10.1093/nar/24.9.1747
[40] LAI C Y, CRONAN J E. Isolation and characterization of beta-ketoacyl-acyl carrier protein reductase (fabG) mutants of Escherichia coli and Salmonella enterica serovar Typhimurium[J]. Journal of Bacteriology, 2004, 186(6): 1869-1878. doi: 10.1128/JB.186.6.1869-1878.2004
[41] WANG H H, CRONAN J E. Only one of the two annotated Lactococcus lactis fabG genes encodes a functional beta-ketoacyl-acyl carrier protein reductase[J]. Biochemistry, 2004, 43(37): 11782-11789. doi: 10.1021/bi0487600
[42] FENG S X, MA J C, YANG J, et al. Ralstonia solanacearum fatty acid composition is determined by interaction of two 3-ketoacyl-acyl carrier protein reductases encoded on separate replicons[J]. BMC Microbiology, 2015, 15: 223.
[43] MAO Y H, LI F, MA J C, et al. Sinorhizobium meliloti functionally replaces 3-oxoacyl-acyl carrier protein reductase (FabG) by overexpressing NodG during fatty acid synthesis[J]. Molecular Plant-Microbe Interactions: MPMI, 2016, 29(6): 458-467. doi: 10.1094/MPMI-07-15-0148-R
[44] HU Z, DONG H J, MA J C, et al. Novel Xanthomonas campestris long-chain-specific 3-oxoacyl-acyl carrier protein reductase involved in diffusible signal factor synthesis[J]. mBio, 2018, 9(3): e00596-e00518.
[45] GUO Q Q, ZHANG W B, ZHANG C, et al. Characterization of 3-oxacyl-acyl carrier protein reductase homolog genes in Pseudomonas aeruginosa PAO1[J]. Frontiers in Microbiology, 2019, 10: 1028.
[46] CUKIER C D, HOPE A G, ELAMIN A A, et al. Discovery of an allosteric inhibitor binding site in 3-oxo-acyl-ACP reductase from Pseudomonas aeruginosa[J]. ACS Chemical Biology, 2013, 8(11): 2518-2527.
[47] JOERNVALL H, PERSSON B, KROOK M, et al. Short-chain dehydrogenases/reductases (SDR)[J]. Biochemistry, 1995, 34(18): 6003-6013. doi: 10.1021/bi00018a001
[48] PERSSON B, KALLBERG Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs)[J]. Chemico Biological Interactions, 2013, 202(1): 111-115.
[49] LORD D M, BARAN A U, WOOD T K, et al. BdcA, a protein important for Escherichia coli biofilm dispersal, is a short-chain dehydrogenase/reductase that binds specifically to NADPH[J]. PLoS One, 2014, 9(9): e105751.
[50] FISHER M, KROON J T, MARTINDALE W, et al. The X-ray structure of Brassica napus β-keto acyl carrier protein reductase and its implications for substrate binding and catalysis[J]. Structure, 2000, 8(4): 339-347. doi: 10.1016/S0969-2126(00)00115-5
[51] BUYSSCHAERT G, VERSTRAETE K, SAVVIDES S N, et al. Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference[J]. The FEBS Journal, 2013, 280(5): 1358-1370. doi: 10.1111/febs.12128
[52] YANG J K, YOON H J, AHN H J, et al. Crystallization and preliminary X-ray crystallographic analysis of the Rv2002 gene product from Mycobacterium tuberculosis, a beta-ketoacyl carrier protein reductase homologue[J]. Acta Crystallographica Section D, Biological Crystallography, 2002, 58(Pt2): 303-305.
[53] REN Q, SIERRO N, WITHOLT B, et al. FabG, an NADPH-dependent 3-ketoacyl reductase of Pseudomonas aeruginosa, provides precursors for medium-chain-length poly-3-hydroxyalkanoate biosynthesis in Escherichia coli[J]. Journal of Bacteriology, 2000, 182(10): 2978-2981. doi: 10.1128/JB.182.10.2978-2981.2000
[54] PRICE A C, ZHANG Y M, ROCK C O, et al. Structure of β-ketoacyl-[acyl carrier protein] reductase from Escherichia coli: Negative cooperativity and its structural asis[J]. Biochemistry, 2001, 40(43): 12772-12781.
[55] CROSS E M, ADAMS F G, WATERS J K, et al. Insights into Acinetobacter baumannii fatty acid synthesis 3-oxoacyl-ACP reductases[J]. Scientific Reports, 2021, 11(1): 1-16. doi: 10.1038/s41598-020-79139-8
[56] VELLA P, RUDRARAJU R S, LUNDBäCK T, et al. A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from gram-negative ESKAPE pathogens[J]. Bioorganic & Medicinal Chemistry, 2021, 30: 115898.
[57] PRICE A C, ZHANG Y M, ROCK C O, et al. Cofactor-induced conformational rearrangements establish a catalytically competent active site and a proton relay conduit in FabG[J]. Structure, 2004, 12(3): 417-428. doi: 10.1016/j.str.2004.02.008
[58] ZHANG Y M, WU B N, ZHENG J, et al. Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction[J]. The Journal of Biological Chemistry, 2003, 278(52): 52935-52943. doi: 10.1074/jbc.M309874200
[59] 童文华, 张文彬, 马金成, 等. 大肠杆菌3−酮基脂酰ACP还原酶110位天冬酰胺突变后的结构与功能[J]. 中国生物化学与分子生物学报, 2012, 28(8): 713-721. doi: 10.13865/j.cnki.cjbmb.2012.08.010 [60] HU Z, MA J C, CHEN Y C, et al. Escherichia coli FabG 3-ketoacyl-ACP reductase proteins lacking the assigned catalytic triad residues are active enzymes[J]. The Journal of Biological Chemistry, 2021, 296: 100365.
[61] CROSS E M, ARAGÃO D, SMITH K M, et al. Structural characterization of a short-chain dehydrogenase/reductase from multi-drug resistant Acinetobacter baumannii[J]. Biochemical and Biophysical Research Communications, 2019, 518(3): 465-471. doi: 10.1016/j.bbrc.2019.08.056
[62] ZHANG Y M, WHITE S W, ROCK C O. Inhibiting bacterial fatty acid synthesis[J]. Journal of Biological Chemistry, 2006, 281(26): 17541-17544. doi: 10.1074/jbc.R600004200
[63] ZHENG Y M, ZHU Y S, MAO X H, et al. SDR7-6, a short-chain alcohol dehydrogenase/reductase family protein, regulates light-dependent cell death and defence responses in rice[J]. Molecular Plant Pathology, 2022, 23(1): 78-91. doi: 10.1111/mpp.13144
[64] ZHANG Y M, ROCK C O. Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase[J]. Journal of Biological Chemistry, 2004, 279(30): 30994-31001. doi: 10.1074/jbc.M403697200
[65] WU D, WU X D, YOU X F, et al. Inhibitory effects on bacterial growth and beta-ketoacyl-ACP reductase by different species of maple leaf extracts and tannic acid[J]. Phytotherapy Research: PTR, 2010, 24(S1): S35-S41. doi: 10.1002/ptr.2873
[66] ENGEN K, ROSENSTRÖM U, AXELSSON H, et al. Identification of drug-Like inhibitors of insulin-regulated aminopeptidase through small-molecule screening[J]. Assay and Drug Development Technologies, 2016, 14(3): 180-193. doi: 10.1089/adt.2016.708
[67] VARAKALA S D, RESHMA R S, SCHNELL R, et al. Lead derivatization of ethyl 6-bromo-2-((dimethylamino) methyl)-5-hydroxy-1-phenyl-1H-indole-3-carboxylate and 5-bromo-2-(thiophene-2-carboxamido) benzoic acid as FabG inhibitors targeting ESKAPE pathogens[J]. European Journal of Medicinal Chemistry, 2022, 228: 113976.
[68] KRISTAN K, BRATKOVIC T, SOVA M, et al. Novel inhibitors of beta-ketoacyl-ACP reductase from Escherichia coli[J]. Chemico Biological Interactions, 2009, 178(1): 310-316.
[69] BANERJEE A, SUGANTINO M, SACCHETTINI J C, et al. The mabA gene from the inhA operon of Mycobacterium tuberculosis encodes a 3-ketoacyl reductase that fails to confer isoniazid resistance[J]. Microbiology, 1998, 144(10): 2697-2704. doi: 10.1099/00221287-144-10-2697
[70] MARRAKCHI H, DUCASSE S, LABESSE G, et al. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II[J]. Microbiology, 2002, 148(Pt4): 951-960.
[71] PARISH T, ROBERTS G, LAVAL F, et al. Functional complementation of the essential gene fabG1 of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG but not Escherichia coli fabG[J]. Journal of Bacteriology, 2007, 189(10): 3721-3728. doi: 10.1128/JB.01740-06
[72] DENG Y Y, WU J E, TAO F, et al. Listening to a new language: DSF-based quorum sensing in gram-negative bacteria[J]. Chemical Reviews, 2011, 111(1): 160-173. doi: 10.1021/cr100354f
[73] 周莲, 王杏雨, 何亚文. 植物病原黄单胞菌DSF信号依赖的群体感应机制及调控网络[J]. 中国农业科学, 2013, 46(14): 2910-2922. doi: 10.3864/j.issn.0578-1752.2013.14.007 [74] YU Y H, MA J R, GUO Q Q, et al. A novel 3-oxoacyl-ACP reductase (FabG3) is involved in the xanthomonadin biosynthesis of Xanthomonas campestris pv. campestris[J]. Molecular Plant Pathology, 2019, 20(12): 1696-1709. doi: 10.1111/mpp.12871
[75] SINGH R, REYNOLDS K A. Characterization of FabG and FabI of the Streptomyces coelicolor dissociated fatty acid synthase[J]. Chembiochem, 2015, 16(4): 631-640. doi: 10.1002/cbic.201402670
-
期刊类型引用(1)
1. 陆钊梅,Kashif Muhammad,莫淑名,杜林方,蒋承建. 铵胁迫下Bacillus aryabhattai NM1-A2脂质代谢的多组学分析. 四川大学学报(自然科学版). 2024(02): 184-191 .  百度学术
百度学术
其他类型引用(0)



 下载:
下载: