Targeting autophagic degradation techniques and potential applications in plant science
-
摘要:
靶向降解技术是一类利用真核细胞内天然存在的降解机制对胞内有害物质进行特异降解、以维持和改善细胞稳态的重要技术。该技术主要通过泛素−蛋白酶体系统 (Ubiquitin-proteasome system,UPS) 和自噬−溶酶体途径 (Autophagy-lysosome pathway),特异性清除细胞内错误折叠或聚集的蛋白质、大分子复合物、受损或老化的细胞器及一些非蛋白类物质。其中基于细胞自噬的靶向降解技术具有专一性强、底物种类广泛等诸多特征,使其成为一备受期待的技术,有望应用于神经退行性疾病、代谢性疾病等多种疾病的治疗。目前这一技术的应用潜能还远未被完全开发,特别是在植物研究领域。本综述首先详细介绍了各类基于自噬−溶酶体途径的靶向降解技术的作用机制、特点以及优势;并且结合华南农业大学李发强教授课题组的研究工作,着重介绍了设计和改造植物选择性自噬衔接蛋白方面的研究和设想,以期达到将对植物生长发育不利的因子经由细胞自噬转运并区室化隔离于液泡的目的,进而开发能够抵御病毒侵染或抵抗有害物质的农作物新品种;最后展望了靶向自噬的降解技术在植物科学研究和农业生产中的潜在应用前景和所面临的挑战。
Abstract:Targeted degradation techniques use the naturally existing degradation mechanism(s) in eukaryotic cells to specifically degrade harmful substances to maintain and improve cellular homeostasis. The techniques specifically remove misfolded or aggregated proteins, macromolecular complexes, damaged or aged organelles, and some non-protein substances via the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway. Among these techniques, the targeting autophagic degradation techniques have many characteristics such as high target selectivity and wide substrate scope, making them promising techniques for the treatment of neurodegenerative diseases, metabolic diseases, and other diseases. At present, the application potential of these techniques is far from being fully developed, especially in the field of plant science. This review details the mechanisms, characteristics, and advantages of targeted degradation techniques based on the autophagy-lysosomal pathway. Moreover, combined with the recent research work of Professor Li Faqiang group in South China Agricultural University, this review focuses on the research of designing and modifying the autophagy adaptors to transport those factors unfavorable to plant growth and development via selective autophagy to the vacuole for compartmentation, so as to develop new crop varieties that can resist viral infection or toxic abiotic components. This review ends with a discussion of the potential applications and challenges of targeted autophagic degradation techniques in plant science and agricultural production.
-
Keywords:
- Targeting degradation technique /
- Autophagy /
- Selective autophagy /
- Autophagy adaptor /
- Vacuole
-
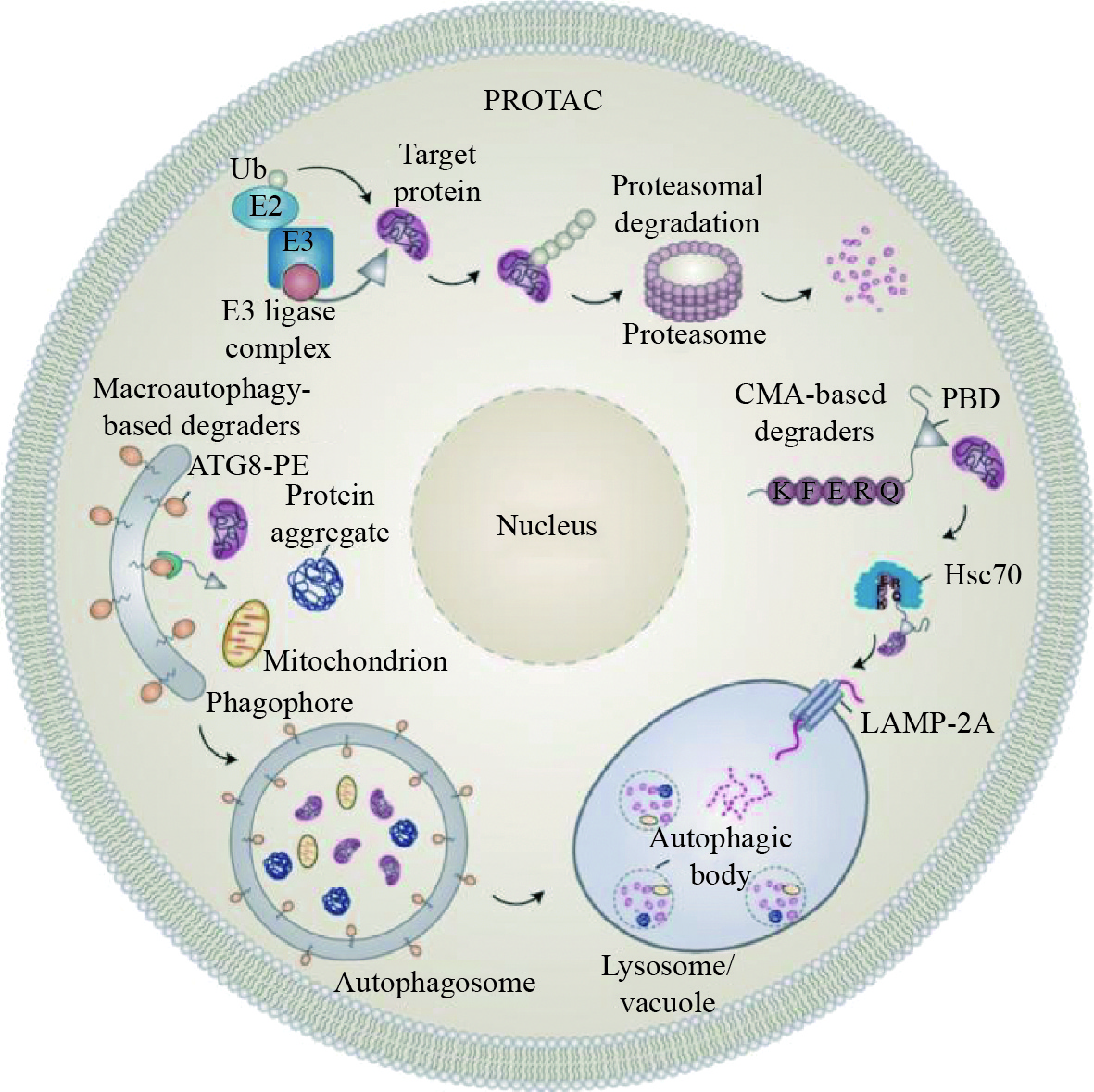
图 1 基于泛素−蛋白酶体系统和自噬−溶酶体/液泡途径的靶向降解技术
依赖于泛素−蛋白酶体系统的蛋白水解靶向嵌合体 (PROTAC) 技术可以通过蛋白酶体降解细胞内的目标蛋白;利用自噬−溶酶体/液泡降解途径的新技术有可能降解胞内蛋白、蛋白聚集体、受损或老化的细胞器、病毒等物质,包括基于巨自噬和基于分子伴侣介导的自噬(CMA) 的靶向降解技术
Figure 1. Targeted degradation techniques based on the ubiquitin-proteasome system and the autophagy-lysosome/vacuole pathway
The proteolysis-targeting chimera (PROTAC) degrades the target proteins via the ubiquitin-proteasome system. Novel techniques based on the autophagy-lysosome/vacuole pathway are anticipated to be applied to remove the target or aggregated proteins, damaged or aged organelles, and viruses, etc. The macroautophagy-based degraders and the chaperone-mediated autophagy (CMA)-based degraders are two of the examples
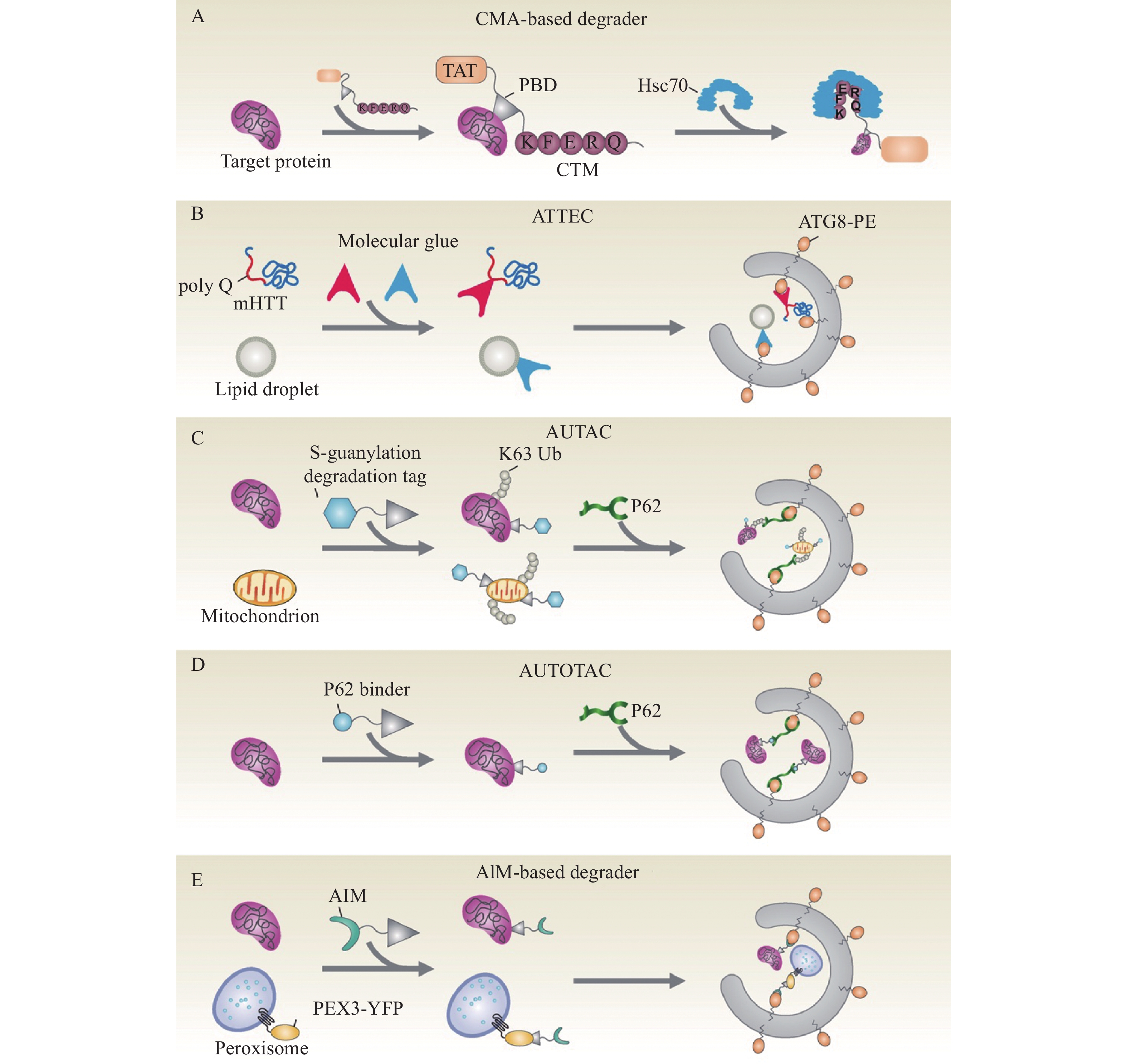
图 2 基于自噬−溶酶体/液泡途径的靶向降解策略示意图
A:基于CMA的降解剂(CMA-based degrader)包括3个功能域——1个细胞膜穿透序列 (TAT)、1个靶向底物的特异配体以及1个CMA靶向模块(CTM);利用TAT进入细胞后,降解剂可以结合底物并诱导伴侣介导的自噬进行特异性降解。B:自噬体绑定化合物ATTECs与LC3/ATG8和靶标底物(mHTT或脂滴) 相互作用,将底物连接到自噬体,以进行随后的自噬降解。C:AUTAC通过靶向底物的特异配体与底物(目标蛋白或线粒体)结合,并利用模仿鸟嘌呤衍生物的降解标签触发靶标底物的K63多泛素化 (Ub),靶标底物随后被自噬受体SQSTM1/p62识别,招募到选择性自噬途径进行降解。D:AUTOTAC分子与靶标底物和p62衔接蛋白相互作用,进而将靶标底物通过自噬途径转运到溶酶体中进行降解。E:基于ATG8互作基序(AIM)的降解剂可以通过与靶标底物和ATG8相互作用,将底物连接到自噬体再进入液泡降解
Figure 2. Schematic diagram for the targeted degradation technique based on the autophagy-lysosome/vacuole pathway
A: The tripartite CMA-based degrader contains the cell membrane-penetrating sequence (TAT) followed by a target protein binding domain (PBD) and tandem repeats of the CMA-targeting motif (CTM); Aided by the TAT, these chimeric proteins enter the cell and bind to the target protein, resulting in specific degradation via CMA. B: The autophagosome-tethering compound (ATTEC) that interacts with both the LC3/ATG8 and the target substrates (mHTT or lipid droplets) targets the latter for autophagic clearance. C: The autophagy-targeting chimera (AUTAC) consists of a specific binder of an intracellular substrate (target protein or mitochondria) and a degradation tag (guanine derivatives) to induce K63-linked ubiquitination of the substrate; The target substrate later is recognized by the autophagy receptor SQSTM1/p62, resulting in autophagic degradation. D: The AUTOphagy-TArgeting Chimera (AUTOTAC) interacting with both the target substrate and the autophagy receptor p62, targets the cargo to lysosomes for degradation. E: The AIM (ATG8-interacting motif)-based degrader interacts with both the target substrate and the ATG8, enclosing the substrate into the autophagosome and later to the vacuole for degradation
-
[1] VERMA R, MOHL D, DESHAIES R J. Harnessing the power of proteolysis for targeted protein inactivation[J]. Molecular Cell, 2020, 77(3): 446-460. doi: 10.1016/j.molcel.2020.01.010
[2] SAKAMOTO K M, KIM K B, KUMAGAI A, et al. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98(15): 8554-8559. doi: 10.1073/pnas.141230798
[3] DANG C V, REDDY E P, SHOKAT K M, et al. Drugging the ‘undruggable’ cancer targets[J]. Nature Reviews Cancer, 2017, 17(8): 502-508. doi: 10.1038/nrc.2017.36
[4] DING Y, FEI Y, LU B. Emerging new concepts of degrader technologies[J]. Trends in Pharmacological Sciences, 2020, 41(7): 464-474. doi: 10.1016/j.tips.2020.04.005
[5] BARD J A M, GOODALL E A, GREENE E R, et al. Structure and function of the 26S proteasome[J]. Annual Review of Biochemistry, 2018, 87: 697-724. doi: 10.1146/annurev-biochem-062917-011931
[6] BURSLEM G M, CREWS C M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery[J]. Cell, 2020, 181(1): 102-114. doi: 10.1016/j.cell.2019.11.031
[7] HINES J, GOUGH J D, CORSON T W, et al. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(22): 8942-8947. doi: 10.1073/pnas.1217206110
[8] REYNDERS M, MATSUURA B S, BEROUTI M, et al. PHOTACs enable optical control of protein degradation[J]. Science Advances, 2020, 6(8): eaay5064. doi: 10.1126/sciadv.aay5064
[9] MARSHALL R S, VIERSTRA R D. Autophagy: The master of bulk and selective recycling[J]. Annual Review of Plant Biology, 2018, 29(69): 173-208.
[10] GALLUZZI L, BAEHRECKE E H, BALLABIO A, et al. Molecular definitions of autophagy and related processes[J]. EMBO Journal, 2017, 36(13): 1811-1836. doi: 10.15252/embj.201796697
[11] LEVINE B, KROEMER G. Biological functions of autophagy genes: A disease perspective[J]. Cell, 2019, 176(1/2): 11-42.
[12] KAUSHIK S, CUERVO A M. Chaperone-mediated autophagy: A unique way to enter the lysosome world[J]. Trends in Cell Biology, 2012, 22(8): 407-417. doi: 10.1016/j.tcb.2012.05.006
[13] BAUER P O, GOSWAMI A, WONG H K, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein[J]. Nature Biotechnology, 2010, 28(3): 256-263. doi: 10.1038/nbt.1608
[14] FAN X, JIN W Y, LU J, et al. Rapid and reversible knockdown of endogenous proteins by peptide-directed lysosomal degradation[J]. Nature Neuroscience, 2014, 17(3): 471-480. doi: 10.1038/nn.3637
[15] WANG H, YAO H, LI C, et al. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity[J]. Nature Chemical Biology, 2019, 15(1): 42-50. doi: 10.1038/s41589-018-0161-x
[16] ZHOU Y F, WANG J, DENG M F, et al. The peptide-directed lysosomal degradation of CDK5 exerts therapeutic effects against stroke[J]. Aging and Disease, 2019, 10(5): 1140-1145. doi: 10.14336/AD.2018.1225
[17] LAMARK T, JOHANSEN T. Mechanisms of selective autophagy[J]. Annual Review of Cell and Developmental Biology, 2021, 37: 143-169. doi: 10.1146/annurev-cellbio-120219-035530
[18] JOHANSEN T, LAMARK T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors[J]. Journal of Molecular Biology, 2020, 432(1): 80-103. doi: 10.1016/j.jmb.2019.07.016
[19] BIRGISDOTTIR A B, LAMARK T, JOHANSEN T. The LIR motif: Crucial for selective autophagy[J]. Journal of Cell Science, 2013, 126(15): 3237-3247. doi: 10.1242/jcs.126128
[20] LEBER R, SILLES E, SANDOVAL I V, et al. Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole[J]. Journal of Biological Chemistry, 2001, 276(31): 29210-29217. doi: 10.1074/jbc.M101438200
[21] SCOTT S V, GUAN J, HUTCHINS M U, et al. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway[J]. Molecular Cell, 2001, 7(6): 1131-1141. doi: 10.1016/S1097-2765(01)00263-5
[22] BJORKOY G, LAMARK T, BRECH A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death[J]. Journal of Cell Biology, 2005, 171(4): 603-614. doi: 10.1083/jcb.200507002
[23] PANKIV S, CLAUSEN T H, LAMARK T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy[J]. Journal of Biological Chemistry, 2007, 282(33): 24131-24145. doi: 10.1074/jbc.M702824200
[24] LUO S, LI X, ZHANG Y, et al. Cargo recognition and function of selective autophagy receptors in plants[J]. International Journal of Molecular Sciences, 2021, 22(3): 1013. doi: 10.3390/ijms22031013.
[25] STEPHANI M, DAGDAS Y. Plant selective autophagy: Still an uncharted territory with a lot of hidden gems[J]. Journal of Molecular Biology, 2020, 432(1): 63-79. doi: 10.1016/j.jmb.2019.06.028
[26] LI Z, WANG C, WANG Z, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds[J]. Nature, 2019, 575(7781): 203-209. doi: 10.1038/s41586-019-1722-1
[27] FU Y, CHEN N, WANG Z, et al. Degradation of lipid droplets by chimeric autophagy-tethering compounds[J]. Cell Research, 2021, 31(9): 965-979. doi: 10.1038/s41422-021-00532-7
[28] TAKAHASHI D, MORIYAMA J, NAKAMURA T, et al. AUTACs: Cargo-specific degraders using selective autophagy[J]. Molecular Cell, 2019, 76(5): 797-810. doi: 10.1016/j.molcel.2019.09.009
[29] ITO C, SAITO Y, NOZAWA T, et al. Endogenous nitrated nucleotide is a key mediator of autophagy and innate defense against bacteria[J]. Molecular Cell, 2013, 52(6): 794-804. doi: 10.1016/j.molcel.2013.10.024
[30] JI C H, KIM H Y, LEE M J, et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system[J]. Nature Communications, 2022, 13(1): 904. doi: 10.1038/s41467-022-28520-4.
[31] LUO N, SHANG D, TANG Z, et al. Engineered AIM-based selective autophagy to degrade proteins and organelles[EB/OL]. BioRxiv: 2021.06.11.448008 (2021-06-11) [2022-08-26]. https://doi.org/10.1101/2021.06.11.448008.
[32] MININA E A, MOSCHOU P N, VETUKURI R R, et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness[J]. Journal of Experimental Botany, 2018, 69(6): 1415-1432. doi: 10.1093/jxb/ery010
[33] ZHEN X, ZHENG N, YU J, et al. Autophagy mediates grain yield and nitrogen stress resistance by modulating nitrogen remobilization in rice[J]. PLoS One, 2021, 16(1): e0244996. doi: 10.1371/journal.pone.0244996
[34] ZHEN X, XU F, ZHANG W, et al. Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis[J]. PLoS One, 2019, 14(9): e0223011. doi: 10.1371/journal.pone.0223011
[35] ZHEN X, LI X, YU J, et al. OsATG8c-mediated increased autophagy regulates the yield and nitrogen use efficiency in rice[J]. International Journal of Molecular Sciences, 2019, 20(19): 4956. doi: 10.3390/ijms20194956.
[36] YU J, ZHEN X, LI X, et al. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE)[J]. Frontiers in Plant Science, 2019, 10: 584. doi: 10.3389/fpls.2019.00584.
[37] CHEN Q, SOULAY F, SAUDEMONT B, et al. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling[J]. Plant and Cell Physiology, 2019, 60(2): 343-352. doi: 10.1093/pcp/pcy214
[38] AVIN-WITTENBERG T, BALUSKA F, BOZHKOV P V, et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection[J]. Journal of Experimental Botany, 2018, 69(6): 1335-1353. doi: 10.1093/jxb/ery069
[39] YANG M, ISMAYIL A, LIU Y. Autophagy in plant-virus interactions[J]. Annual Review of Virology, 2020, 7: 403-419. doi: 10.1146/annurev-virology-010220-054709
[40] HAFREN A, USTUN S, HOCHMUTH A, et al. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro[J]. Plant Physiology, 2018, 176(1): 649-662. doi: 10.1104/pp.17.01198
[41] LI F, ZHANG C, LI Y, et al. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase[J]. Nature Communications, 2018, 9: 1268. doi: 10.1038/s41467-018-03658-2.
[42] HAXIM Y, ISMAYIL A, JIA Q, et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants[J]. eLife, 2017, 6: e23897. doi: 10.7554/eLife.23897
[43] JIANG L, LU Y, ZHENG X, et al. The plant protein NbP3IP directs degradation of rice stripe virus p3 silencing suppressor protein to limit virus infection through interaction with the autophagy-related protein NbATG8[J]. New Phytologist, 2021, 229(2): 1036-1051. doi: 10.1111/nph.16917
[44] NAKAHARA K S, MASUTA C, YAMADA S, et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(25): 10113-10118. doi: 10.1073/pnas.1201628109
[45] MICHAELI S, CLAVEL M, LECHNER E, et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(45): 22872-22883. doi: 10.1073/pnas.1912222116
[46] GHANNAM A, KUMARI S, MUYLDERMANS S, et al. Camelid nanobodies with high affinity for broad bean mottle virus: A possible promising tool to immunomodulate plant resistance against viruses[J]. Plant Molecular Biology, 2015, 87(4/5): 355-369.
[47] HEMMER C, DJENNANE S, ACKERER L, et al. Nanobody-mediated resistance to Grapevine fanleaf virus in plants[J]. Plant Biotechnology Journal, 2018, 16(2): 660-671. doi: 10.1111/pbi.12819
[48] YANG B, YANG S, ZHENG W, et al. Plant immunity inducers: From discovery to agricultural application[J/OL]. Stress Biology, 2022, 2: 5. (2022-01-10) [2022-09-24]. https://doi.org/10.1007/s44154-021-00028-9.
[49] WANG J W, GRANDIO E G, NEWKIRK G M, et al. Nanoparticle-mediated genetic engineering of plants[J]. Molecular Plant, 2019, 12(8): 1037-1040.
[50] CLEMENS S, MA J F. Toxic heavy metal and metalloid accumulation in crop plants and foods[J]. Annual Review of Plant Biology, 2016, 67: 489-512. doi: 10.1146/annurev-arplant-043015-112301
[51] SASAKI A, YAMAJI N, YOKOSHO K, et al. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice[J]. Plant Cell, 2012, 24(5): 2155-2167. doi: 10.1105/tpc.112.096925
[52] UENO D, KONO I, YOKOSHO K, et al. A major quantitative trait locus controlling cadmium translocation in rice (Oryza sativa)[J]. New Phytologist, 2009, 182(3): 644-653. doi: 10.1111/j.1469-8137.2009.02784.x
[53] MIYADATE H, ADACHI S, HIRAIZUMI A, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles[J]. New Phytologist, 2011, 189(1): 190-199. doi: 10.1111/j.1469-8137.2010.03459.x
[54] SATOH-NAGASAWA N, MORI M, NAKAZAWA N, et al. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium[J]. Plant and Cell Physiology, 2012, 53(1): 213-224. doi: 10.1093/pcp/pcr166
[55] TAKAHASHI R, ISHIMARU Y, SHIMO H, et al. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice[J]. Plant Cell and Environment, 2012, 35(11): 1948-1957. doi: 10.1111/j.1365-3040.2012.02527.x
[56] URAGUCHI S, KAMIYA T, SAKAMOTO T, et al. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(52): 20959-20964. doi: 10.1073/pnas.1116531109
[57] LUO J S, HUANG J, ZENG D L, et al. A defensin-like protein drives cadmium efflux and allocation in rice[J]. Nature Communications, 2018, 9: 645. doi: 10.1038/s41467-018-03088-0.
[58] PENG J S, DING G, MENG S, et al. Enhanced metal tolerance correlates with heterotypic variation in SpMTL, a metallothionein-like protein from the hyperaccumulator Sedum plumbizincicola[J]. Plant, Cell and Environment, 2017, 40(8): 1368-1378. doi: 10.1111/pce.12929



 下载:
下载: