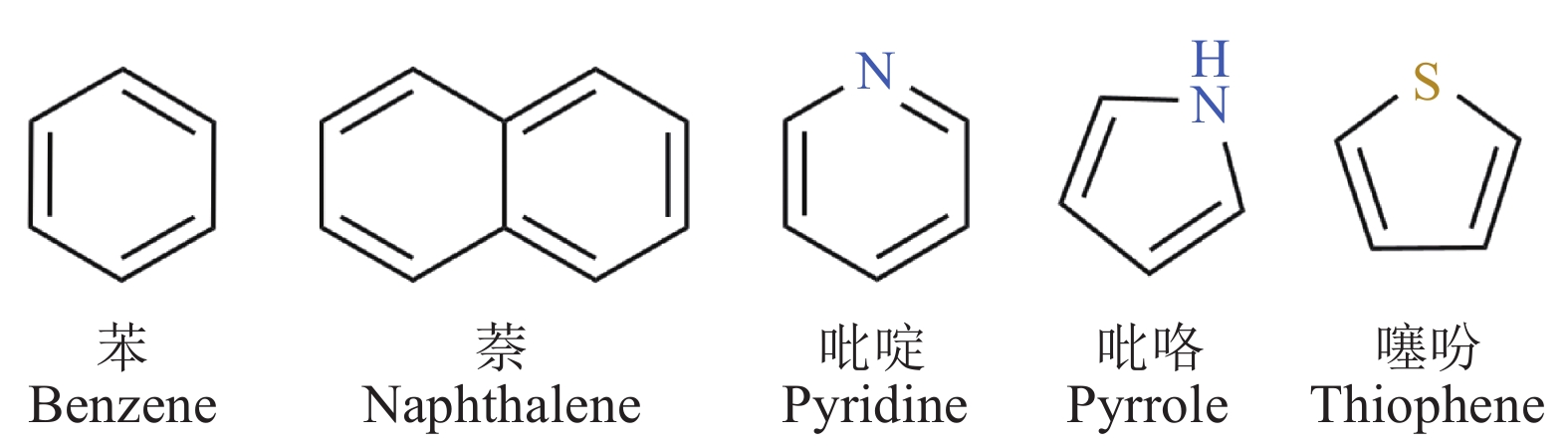
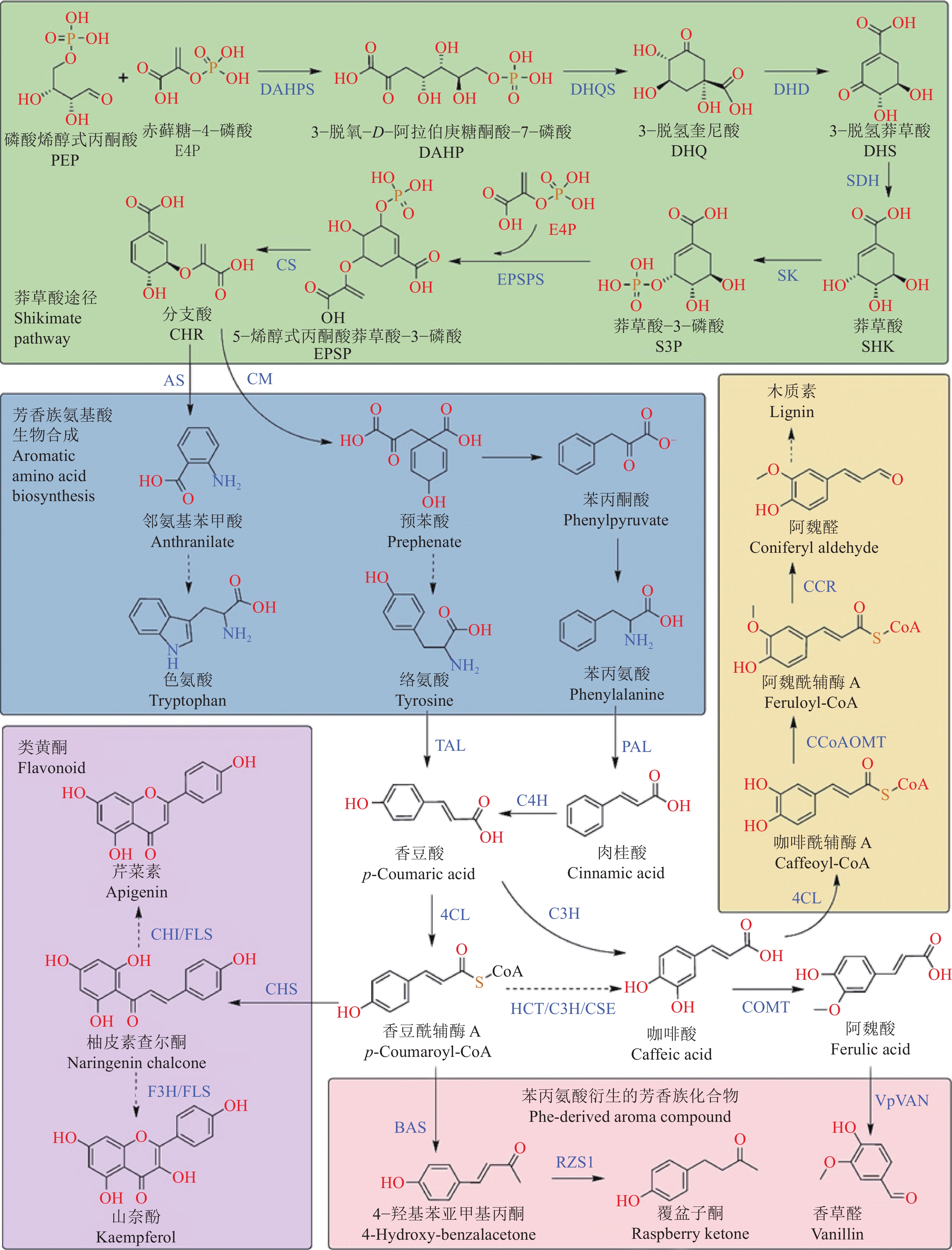
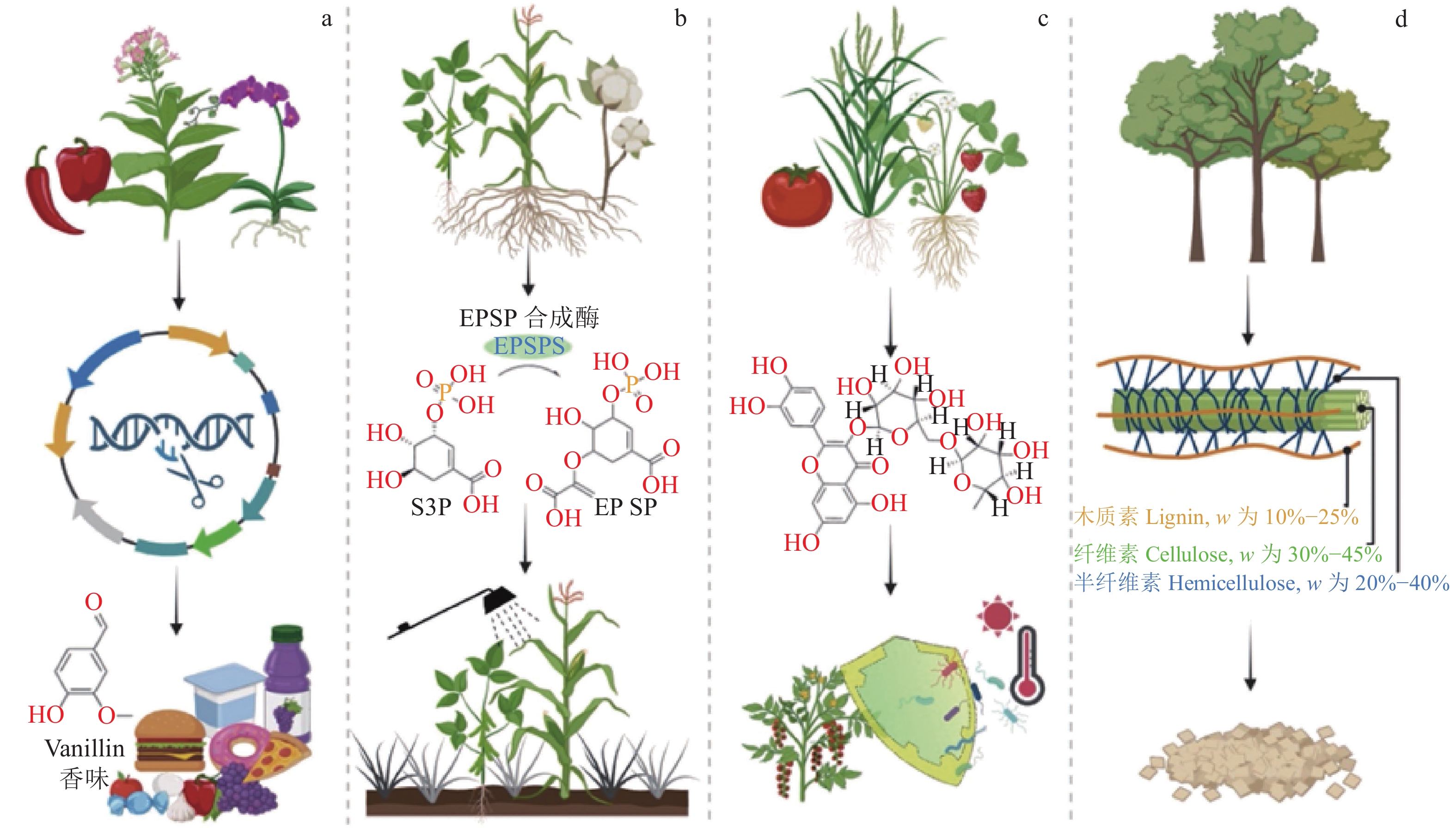
Research progress on synthetic biology of aromatic compounds and their application in biological breeding
-
摘要:
芳香族化合物是一类含有芳香环结构的有机小分子,主要产生于植物和微生物,也可以化学合成,在化学、材料和生命科学等领域具有重要的应用价值。本文介绍了芳香族化合物的生物合成途径和合成生物学方法,综述了其在生物育种中提高植物的香味、类黄酮和培育抗除草剂、低木质素种质等方面的应用,并对合成生物学在芳香族化合物研究中的未来前景进行了展望。
Abstract:Aromatic compounds are organic small molecules that contain one or more aromatic rings in their structures. They are mainly produced by plants and microorganisms, but can also be synthesized chemically. Aromatic compounds have important applications in chemistry, materials and life sciences. This review summarized the biosynthesis pathways and synthetic biology of aromatic compounds, as well as their potential uses in improving the aroma, flavonoid content, herbicide tolerance, and lignin reduction of plants through biotechnology. The future prospect of synthetic biology in the research of aromatic compounds was discussed.
-
近年来,梨小食心虫Grapholitha molesta已经成为我国很多果区的重要害虫,在桃、梨和苹果园造成严重危害,并有不断上升的趋势,国内主要分布在东北、华北、西北、华东等地区的省市,国外分布也比较广泛,主要发生于亚、欧、南美和北美洲,目前已成为世界性蛀果害虫[1-5]。梨小食心虫在早春以蛀食桃或苹果新梢为主,后期转移为害各种果实,尤其在桃树与梨树的混栽区,危害更为严重。长期使用化学农药防控梨小食心虫可造成环境污染,且害虫抗药性增强,而昆虫性信息素作为一种新型绿色防控技术,不仅可以用于监测害虫的发生动态,还可以大量诱捕害虫,尤其迷向干扰技术可用于大面积防控害虫,在害虫综合防治中起着非常重要的作用[6-8],但是由于人工合成的昆虫性信息素大多数为挥发性化合物,野外应用中易受外界环境的影响,尤其是持效期短,严重制约着昆虫性信息素技术的大面积推广应用,要使性信息素在野外使用中长期保持一定浓度而且成分比例相对稳定的状态,有必要使用微胶囊缓释技术[9-10],延长性信息素的作用时间及控制性信息素的释放速度,进而应用性信息素微胶囊乳液迷向干扰技术防治害虫[11]。
微胶囊缓释包埋工艺可显著延长挥发性化合物的持效时间,它通过高分子材料把固体、液体或气体包埋并贮存在一种具有半透性或密封性囊膜的微型胶囊内,制备成一种微粒 [12-16],其粒径范围为5~200 μm,囊壁厚度范围为0.2~10 μm,包埋在微胶囊内部的材料称为芯材,包附在芯材外的材料称为壁材。因此,昆虫性信息素经微胶囊包埋工艺处理后,在包膜内可形成微胶囊,进而转化为稳定的水溶性溶液,通过包膜缓慢释放到野外,促使林间持续弥漫性信息素气味,大大降低雌雄成虫的交配率。
目前,市场上售卖的梨小食心虫防治性信息素类药物基本都是以悬挂“性信息素迷向丝”干扰雌雄交配[17-18],例如:吴甚妍等[19]报道了迷向丝用于黄桃园梨小食心虫的防控研究,对2、3代成虫迷向效果显著;郑鹏华等[20]研究了梨小食心虫性信息素迷向丝在浙北桃园的应用效果对比,在45 d内,迷向率由69.2%提升至100%,并保持稳定;曹敏等[21]利用性信息素迷向瓶和迷向丝研究了新疆蟠桃园梨小食心虫的防治效果,其迷向率分别为84.44%和70.02%;孙圣杰等[22]报道了梨小食心虫迷向散发器可有效干扰害虫的正常交尾,全年范围内降低发生数量。但微胶囊乳液迷向干扰防控梨小食心虫的报道甚少。本文通过微胶囊缓释包埋技术将梨小食心虫性信息素制备成微胶囊乳液,主要通过探索微胶囊乳液的制备过程、缓释时期及野外迷向效果,明确壁材与芯材比例、剪切速度、均质压力及液体石蜡加入量对微胶囊乳液的影响,为进一步利用昆虫性信息素迷向防控梨小食心虫提供可靠的技术支撑。
1. 材料与方法
1.1 试剂和仪器
试验试剂:梨小食心虫性信息素(北京中捷四方公司);β−环糊精、麦芽糊精(上海源聚生物科技有限公司);辛烯基琥珀酸淀粉钠(广州华汇生物实业有限公司);乳化变性淀粉−809(广州艾辉生物科技有限公司);液体石蜡(天津市富宇精细化工有限公司)。
试验仪器:高压均质机(ATS公司),电子天平(梅特勒−托利多仪器有限公司),激光粒度分析仪(英国Malvern仪器有限公司),高剪切乳化均质机(上海标本模型厂),气相色谱仪(Gas chromatography,GC,7820A,Agilent),气相色谱−质谱联用仪(GC-Mass spectrometry,GC-MS,7890A-5975C,Agilent)。
1.2 试验方法
1.2.1 梨小食心虫性信息素微胶囊乳液制备的不同条件选择
壁材溶液的制备:称量20 g试验所需要的不同比例的辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精和β−环糊精,置于150 mL锥形瓶中,加入100 mL纯净水,启动带有大号剪切头的搅拌器,以转速8000~9000 r/min搅拌,直至样品全部溶解。
微胶囊乳液的配制:根据计算,称量试验所需要的梨小食心虫性信息素,慢慢加入到上述制备好的壁材溶液中,启动高剪切乳化均质机以转速10 000 r/min搅拌2 min,直至混合均匀。
微胶囊乳液的高压均质成型:首先用φ为70%的乙醇溶液把仪器内部冲洗洁净,再用纯净水冲洗2~3次,随后顺时针慢速旋转均质阀,等待压力变为稳定值且漏斗内的水到达底部,加入需要均质的样品,出口位置以150 mL锥形瓶承接均质后的样品溶液,均质好的溶液再次加入漏斗中,重复以上步骤4次,再加入液体石蜡(0.4倍壁材用量),充分混匀,并重复以上步骤,待到溶液表面不再出现油状物,达到均质要求。
壁材不同比例筛选: 选择辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精和β−环糊精作为壁材料,将其质量配比依次设置成15∶5∶0∶3∶2、10∶10∶0∶3∶2、5∶15∶0∶3∶2、35∶0∶0∶0∶0、0∶35∶0∶0∶0、0∶0∶20∶3∶2和0∶0∶35∶0∶0共7组处理,梨小食心虫性信息素作为芯材,壁材与芯材质量配比设置成10∶1,剪切速度设置成10 000 r/min,高压均质压力设置成20 MPa,均质持续2 min,常温静置24 h。
不同壁材与芯材比例筛选: 选择辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精和β−环糊精作为壁材料(质量比5∶15∶0∶3∶2),壁材与芯材质量配比依次设置成5∶1、10∶1和40∶1共3组处理,其余设置同“壁材不同比例筛选”。
不同剪切速度筛选: 剪切速度依次设置成5 000、10 000和15 000 r/min,壁材与芯材质量比为10∶1,其余设置同“不同壁材与芯材比例筛选”。
不同均质压力筛选:高压均质压力依次设置成5、20和45 MPa,剪切速度10 000 r/min,其余设置同“不同剪切速度筛选”。
液体石蜡对乳液的影响:壁材与芯材质量比为10∶1、剪切速度10 000 r/min、均质压力20 MPa、均质2 min,然后设置加入液体石蜡0 g (对照)和10 g共2组处理,进行二次均质,均质压力20 MPa、均质持续4 min,加入液体石蜡时进行手动搅拌以使液体石蜡与乳液混合均匀,待乳液表面不再出现油状物时即制备成微胶囊乳液,常温静置24 h。
冷藏时长对乳液的影响:利用上述筛选出的最优条件配制微胶囊乳液。1号样品置于常温24 h,2号样品置于5~10 ℃冷藏2个月,观察记录微胶囊乳液表观情况。
1.2.2 微胶囊乳液的表观观察和粒径测定
配制好的微胶囊乳液常温下静置24 h,转至1 000 mL烧杯中,运用激光粒度分布仪对微胶囊乳液的平均粒径进行测定。
在1 000 mL烧杯中倒入800~900 mL纯净水作为分散剂对微胶囊乳液样品进行分散,搅拌器以2 200 r/min搅拌,运用的乳液分析模式为:颗粒折射率1.467,粒径检测跨度0.02~2 000 μm。检测完背景后,利用一次性胶头滴管吸取梨小食心虫性信息素微胶囊乳液逐滴滴入烧杯内,直至符合检测范围。每一个样品测定完毕后,立即用自来水冲洗至激光背景高于73%。
使用的粒径分布评价指标为D10、D50、D90和跨距[(D90−D10)/D50],其中,D10:小于此粒径的颗粒数占总量的10%;D50:也叫中粒径或中值粒径,小于和大于此粒径的颗粒数均占总量的50%;D90:小于此粒径的颗粒数占总量的90%。D50越小,表示粒径越小;跨距越小,表示粒径分布越集中。一般情况下,常用D50和跨距的均值来评价粒径的分布[23-24]。
1.2.3 微胶囊乳液包封率的测定
精密称量0.100 g梨小食心虫性信息素标准化合物,使用环己烷将其均匀溶解并定容至100 mL。依次吸取0、0.5、1.0、1.5、2.0、2.5和3.0 mL上述溶液于10 mL比色管中,再加入环己烷至刻度,依次得到质量浓度为0、0.05、0.10、0.15、0.20、0.25和0.30 mg/mL的溶液,对溶液进行气相色谱测定,浓度设置成纵坐标,峰面积设置成横坐标,制作标准曲线。
使用移液枪吸取1 mL微胶囊乳液原液加入到2 mL环己烷溶液中,静置均匀后吸取1 μL上层环己烷溶液进行测定,应用气相色谱法测定其表面含量,得微胶囊粒剂中未包封的芯材含量;再用移液枪吸取1 mL微胶囊乳液原液加入到2 mL环己烷溶液中,超声波运行1 h至微胶囊中的芯材物质全部溶解到溶液内,吸取1 μL上层环己烷溶液进行分析,采用气相色谱法检测含量,得微胶囊粒剂原料中芯材含量。气相色谱条件为:HP-5 MS色谱柱(30 m×250 μm,0.25 μm),进样口温度230 ℃,分流比10∶1;检测器(FID)温度250 ℃;升温程序:初始温度150 ℃,20 ℃/min升到190 ℃,再以5 ℃/min升到220 ℃,保持0 min,最后以30 ℃/min升到250 ℃,保持2 min;氮气、氢气和空气的流量分别设置成 25、30和300 mL/min;进样量1 μL。
微胶囊包封率=(原料中芯材含量−未包封的芯材含量)/原料中芯材含量×100%。
1.2.4 微胶囊乳液释放速率的测定
梨小食心虫性信息素稀释液释放速率的测定:使用环己烷作为溶剂,配制约25 mg/mL梨小食心虫性信息素溶液,用进样针吸取1 μL于锡纸上静置,在20~25 ℃室温下进行缓释试验,使用GC-MS测定微胶囊中芯材物质的含量。
微胶囊乳液释放速率的测定:根据筛选出的最优条件配制微胶囊乳液,吸取1 μL配制的微胶囊乳液原液至直径为0.5 cm的圆形锡纸中部,分别置于30、40和50 ℃的烘箱中,定期取样,应用GC-MS测定微胶囊中芯材物质的含量。
GC-MS程序:载气为He,进样口温度为250 ℃,设置为不分流,色谱柱和升温程序同“1.2.3”。
1.2.5 野外迷向试验
野外试验地位于新疆库尔勒沙依东园艺场果园,占地面积约20 hm2,主栽品种为香梨,行距5 m,株距3 m,树龄10年,树高2~3 m,树势整齐。
野外试验共设4个不同处理:处理1和2分别为45和75 g/hm2(性信息素的含量,而非微胶囊乳液的含量);处理3为120 g/hm2(微胶囊乳液为工厂批量生产,北京中捷四方生物科技股份有限公司提供);处理4为75 g/hm2(冷藏1年的性信息素微胶囊乳液);以0 g/hm2空白处理为对照(CK)。每个处理3次重复,共15个小区,每个小区面积1 hm2,小区间设50 m隔离带,隔离带以45 g/ hm2性信息素处理,每个小区悬挂3个三角形诱捕器,诱捕器呈三角形排列,相邻诱捕器之间距离约为30 m,诱捕器挂于树干1.5~2.0 m处,诱芯每月更换1次,每隔5 d调查1次诱蛾量,试验期间园内果树正常管理。
迷向处理:用毛刷把一定量的微胶囊乳液均匀涂抹于每株梨树的主干上,涂抹处距离地面1~2 m。进行迷向处理后,每隔5 d调查记录1次各处理区内诱捕器所诱梨小食心虫数量,调查时间段为4月30日—8月18日(累计111 d)。统计调查结果,计算梨小食心虫性信息素微胶囊乳液对梨小食心虫的迷向率。迷向率=(1−迷向区诱蛾总量/对照区诱蛾总量)×100%。
1.3 数据处理
试验数据采用SPSS软件进行处理,采用ANOVA 进行方差分析,Tukey HSD进行差异显著性检验。
2. 结果与分析
2.1 壁材比例对微胶囊乳液粒径分布及包封率的影响
表1显示,6个样品的包封率都高于99%,4号样品表面存在油状物(芯材物质)。1~3号样品的包封率随着辛烯基琥珀酸淀粉钠比例的降低,呈现出先下降后上升的趋势:1号(99.60%)>2号(99.49%)<3号(99.63%)。从图1可以看出,1~3号样品都存在不同程度的拖尾情况,大颗粒有小峰拖尾,1号样品的拖尾峰大于2号和3号样品,1号样品的跨距和平均粒径都大于2号和3号样品。4~7号样品的粒径都呈现为正态分布,其中,4、5、7号样品的壁材都是单一样品,除了4号样品,5、6、7号样品的包封率均为99.7%左右,但这4个样品的表观显示均为相对黏稠,不便于野外应用,若是加水稀释则其表观特征会发生很大改变。1~6号样品的D10彼此之间均呈现出显著性差异,3号与7号样品没有显著性差异,7个样品的D50、D90彼此之间也呈现出显著性差异(P<0.05)。根据表观分层、包封率及粒径分布的综合情况可以得出,3号样品在各方面的性能表现最为优良。
表 1 壁材比例对微胶囊乳液粒径分布及包封率的影响1)Table 1. Effect of different proportion of wall materials on particle size distribution and encapsulation rate of microcapsule emulsion样品编号
No. of
sample壁材质量比2)
Mass ratios of
wall materials微胶囊乳液表观
Microcapsule emulsion
appearance粒径/μm
Particle diameter跨距
Span包封率/%
Encapsulation
rateD10 D50 D90 1 15∶5∶0∶3∶2 乳白色,不分层 0.304±0.002a 0.733±0.005a 5.857±0.119a 7.581±0.105a 99.60±0.02b 2 10∶10∶0∶3∶2 乳白色,不分层 0.234±0.014c 0.610±0.003b 1.765±0.157b 2.467±0.287bc 99.49±0.02c 3 5∶15∶0∶3∶2 乳白色,不分层 0.136±0.006f 0.535±0.005c 1.569±0.052c 2.672±0.130b 99.63±0.03b 4 35∶0∶0∶0∶0 浅乳白色,不分层,较
黏稠,表面有油状物0.221±0.00d 0.497±0.001e 1.427±0.002d 2.424±0.002c 98.37±0.02d 5 0∶35∶0∶0∶0 浅乳白色,不分层,
较黏稠
0.165±0.001e 0.359±0.000f 0.798±0.001f 1.763±0.003d 99.72±0.02a 6 0∶0∶20∶3∶2 乳黄色,不分层,黏稠 0.257±0.001b 0.518±0.000d 1.037±0.005e 1.504±0.008e 99.75±0.03a 7 0∶0∶35∶0∶0 乳黄色,不分层,黏稠 0.133±0.00f 0.295±0.000g 0.580±0.000g 1.513±0.001e 99.71±0.03a 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD);2)壁材成分包括辛烯基琥珀酸淀粉钠、乳化变性淀粉−809,明胶,麦芽糊精,β−环糊精
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD); 2)The wall material composition includes octenylsuccinate starch sodium, emulsifying starch-809, gelatin, maltodextrin, β-cyclodextrin![]() 图 1 壁材比例对微胶囊乳液粒径分布的影响1~7号样品的辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精、β−环糊精质量比分别为15∶5∶0∶3∶2、10∶10∶0∶3∶2、5∶15∶0∶3∶2、35∶0∶0∶0∶0、0∶35∶0∶0∶0、0∶0∶20∶3∶2和0∶0∶35∶0∶0Figure 1. Effect of different proportion of wall materials on particle size distribution of microcapsule emulsionThe mass ratios of octenylsuccinate starch sodium, emulsifying starch-809, gelatin, maltodextrin, β-cyclodextrin in samples of 1–7 are 15∶5∶0∶3∶2, 10∶10∶0∶3∶2, 5∶15∶0∶3∶2, 35∶0∶0∶0∶0, 0∶35∶0∶0∶0, 0∶0∶20∶3∶2 and 0∶0∶35∶0∶0, respectively
图 1 壁材比例对微胶囊乳液粒径分布的影响1~7号样品的辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精、β−环糊精质量比分别为15∶5∶0∶3∶2、10∶10∶0∶3∶2、5∶15∶0∶3∶2、35∶0∶0∶0∶0、0∶35∶0∶0∶0、0∶0∶20∶3∶2和0∶0∶35∶0∶0Figure 1. Effect of different proportion of wall materials on particle size distribution of microcapsule emulsionThe mass ratios of octenylsuccinate starch sodium, emulsifying starch-809, gelatin, maltodextrin, β-cyclodextrin in samples of 1–7 are 15∶5∶0∶3∶2, 10∶10∶0∶3∶2, 5∶15∶0∶3∶2, 35∶0∶0∶0∶0, 0∶35∶0∶0∶0, 0∶0∶20∶3∶2 and 0∶0∶35∶0∶0, respectively2.2 壁材与芯材比例对微胶囊乳液粒径分布及包封率的影响
从表2可以看出,3个样品的平均粒径随着壁材含量的增大(芯材不变)而逐渐减小,而包封率则逐渐增大。从图2可以看出,3个样品的粒径分布的主峰形态大致相似,主峰后部均存在不同程度的拖尾小峰,其中以2号样品最为明显。3个样品的D10、D50、D90粒径都存在显著性差异(P<0.05)。综合考虑表观分层、包封率及粒径分布,2号样品在各个方面的表现最为良好。
表 2 壁材与芯材比例对微胶囊乳液粒径分布及包封率的影响1)Table 2. Effect of different mass ratios of wall material to core material on particle size distribution and encapsulation rate of microcapsule emulsion样品编号
No. of sample
壁材、芯材质量比
Mass ratio of
wall material to
core material微胶囊乳液表观
Microcapsule
emulsion
appearance粒径/μm
Particle diameter跨距
Span包封率/%
Encapsulation rateD10 D50 D90 1 5∶1 乳白色,不明显分层,上层较下层颜色白 0.294±0.006a 0.673±0.002a 1.528±0.067b 1.825±0.102b 98.93±0.02c 2 10∶1 乳白色,不分层 0.234±0.014b 0.610±0.003b 1.765±0.157a 2.467±0.287a 99.52±0.03b 3 40∶1 乳白色,不分层,黏稠 0.190±0.004c 0.490±0.001c 1.143±0.015c 1.942±0.028b 99.75±0.02a 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD)
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD)2.3 剪切速度对微胶囊乳液粒径分布及包封率的影响
从表3可以看出,随着剪切速度的增加,粒径D50减小,包封率先变大后变小,跨距为1号>3号>2号。从图3可以看出,1号样品存在3个峰,颗粒粒径分布相对分散, 2号存在1个主要的峰,3号大致存在3个峰。3个样品微胶囊乳液表观都未呈现出分层情况,其粒径的D10、D50、D90及跨距都存在显著性差异(P<0.05)。综合考虑表观分层、包封率及粒径分布,2号样品在各个方面表现出的性能最为良好。
表 3 剪切速度对微胶囊乳液粒径分布及包封率的影响1)Table 3. Effect of different shear velocity on particle size distribution and encapsulation rate of microcapsule emulsion样品编号
No. of sample剪切速度/
( r·min−1)
Shear velocity微胶囊乳液表观Microcapsule emulsion appearance 粒径/μm
Particle diameter跨距
Span包封率/%
Encapsulation rateD10 D50 D90 1 5 000 乳白色,不分层 0.837±0.019b 4.832±0.317a 19.088±2.074a 3.708±0.182a 99.40±0.02b 2 10 000 乳白色,不分层 0.891±0.007a 2.228±0.001b 5.334±0.041b 1.994±0.021c 99.55±0.02a 3 15 000 乳白色,不分层 0.115±0.004c 0.964±0.020c 2.621±0.054c 2.598±0.114b 99.22±0.02c 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD)
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD)2.4 均质压力对微胶囊乳液粒径分布及包封率的影响
从表4可以看出,随着均质压力的增大,D50粒径逐渐减小,跨距先减小后增大,包封率则逐渐变大:1号(99.24%)<2号(99.54%)<3号(97.67%)。从图4可以看出,各样品粒径分布都存在一定的拖尾峰,3号样品的拖尾峰相对明显,2号次之,1号最小。3个样品均呈现乳白色,不出现分层情况。1号与2号、2号与3号样品的D10彼此间存在显著性差异,3个样品的D50、D90彼此之间有显著性差异(P<0.05)。综合分析表观分层、包封率及粒径分布,2号样品在各个方面的表现最为良好。
表 4 均质压力对微胶囊乳液粒径分布及包封率的影响1)Table 4. Effect of different homogeneous pressure on particle size distribution and encapsulation rate of microcapsule emulsion样品编号
No. of
sample均质压力/MPa
Homogenization pressure微胶囊乳液表观
Microcapsule emulsion appearance粒径/μm
Particle diameter跨距
Span包封率/% Encapsulation rate D10 D50 D90 1 5 乳白色,不分层 0.202±0.007b 0.718±0.001a 1.936±0.067b 2.410±0.107b 99.24±0.01c 2 20 乳白色,不分层 0.237±0.013a 0.643±0.002b 1.679±0.092c 2.228±0.168b 99.54±0.02b 3 45 乳白色,不分层 0.217±0.011b 0.613±0.000c 3.633±0.213a 5.602±0.364a 99.67±0.02a 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD)
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD)2.5 加入液体石蜡二次均质对微胶囊乳液粒径分布及包封率的影响
从表5可以看出,加入液体石蜡进行二次均质的样品,微胶囊乳液D10与D50变小,跨距与包封率增大,乳液均匀稳定,粒径分布图(图5)不存在明显的拖尾现象,2个样品的D10、D50、D90及跨距均有显著差异(P<0.05)。综合分析表观分层、包封率及粒径分布,加入液体石蜡进行二次均质的样品在各个方面的性能均会增强。
表 5 加入液体石蜡二次均质对微胶囊乳液粒径分布及包封率的影响1)Table 5. Effect of the second homogeneity with liquid paraffin on particle size distribution and encapsulation rate of microcapsule emulsion液体石蜡加入量/g
Added mass of
liquid paraffin微胶囊乳液表观Microcapsule emulsion appearance 粒径/μm
Particle diameter跨距
Span包封率/%
Encapsulation rateD10 D50 D90 0 乳白色,不分层 0.234±0.014a 0.610±0.157a 0.610±0.003b 2.467±0.287b 99.52±0.04b 10 乳白色,不分层 0.067±0.000b 0.138±0.004b 0.688±0.028a 4.564±0.082a 99.61±0.01a 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD)
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD)2.6 冷藏对微胶囊乳液粒径分布及包封率的影响
从表6可以看出,经过2个月的冷藏,样品在表观上会变黏稠,D10、D50和D90都增大,而包封率低于新配制的样品,粒径分布图(图6)峰形比新配制的样品宽而低,表明经过冷藏,微胶囊乳液发生了聚合现象,致使颗粒变大。2个样品的D10、D50和D90都存在显著性差异(P<0.05)。
表 6 冷藏微胶囊乳液粒径分布及包封率的变化1)Table 6. Effect of refrigeration on particle size distribution and encapsulation rate of microcapsule emulsiont冷藏/月
Refrigeration time微胶囊乳液表观
Microcapsule emulsion
appearance粒径/μm
Particle diameter跨距
Span包封率/%
Encapsulation rateD10 D50 D90 0 乳白色,不分层 0.067±0.00b 0.138±0.004b 0.688±0.028b 4.564±0.082a 99.63±0.02a 2 乳白色,不分层,黏稠 0.121±0.001a 0.328±0.001a 1.139±0.003a 3.098±0.011b 99.51±0.04b 1)同列数据后的不同小写字母表示差异显著(P<0.05,Tukey HSD)
1)Different lowercase letters in the same column indicate significant difference (P<0.05, Tukey HSD)2.7 微胶囊乳液室内缓释试验分析
从图7梨小食心虫性信息素稀释液的释放曲线可以看出,性信息素按照固定的速度释放,在5 h左右释放完毕。
从图8可以看出,放置于30 ℃条件下的性信息素剩余量在前70 d高于放置于40 ℃条件下的剩余量,在70 d之后其剩余量与放置于40 ℃条件下的大致相同;放置于40 ℃条件下的剩余量在前42 d高于40 ℃(J)(使用剪切均质未使用高压均质的样品,放置于40 ℃条件下),从42 d至77 d其剩余量与40 ℃(J)大致相同,40 ℃(J)的剩余量在77 d后大于放置于30和40 ℃条件下的剩余量;放置于40 ℃(J)和50 ℃的2个样品的性信息素释放曲线趋势类似,释放曲线相对平缓,放置于50 ℃条件下样品的性信息素剩余量始终维持在比较低的水平;随着缓释时间的延长,4个样品性信息素的剩余量在91 d后近似相同。
2.8 微胶囊乳液野外迷向效果
图9 迷向率曲线显示,迷向处理77 d之前,除了22 d时处理1(迷向率–33.33%)和处理3(迷向率66.67%)外,其他处理的迷向率都高于81%(迷向率为0的无参考意义),80 d之后各处理的迷向率开始降低,87 d之后有个别处理表现出负的迷向率,但从各处理迷向率的趋势能够看出,微胶囊乳液药效可以维持80 d左右。
3. 讨论与结论
根据上述微胶囊乳液配制的试验结果,得出最佳的配制条件为:辛烯基琥珀酸淀粉钠、乳化变性淀粉−809、明胶、麦芽糊精、β−环糊精的质量比为5∶15∶0∶3∶2,壁材、芯材的质量比为10∶1,剪切速度10000 r/min,剪切时间2 min,高压均质压力20 MPa,均质2 min,二次均质加入10 g液体石蜡,均质压力20 MPa、均质时间4 min。以最佳配方配制梨小食心虫微胶囊乳液,室内缓释试验结果表明,梨小食心虫性信息素微胶囊缓释液在室温下放置5 h后释放完毕,而不同温度下的微胶囊乳液释放率不同,30 ℃条件下微胶囊乳液的释放量相对较小,微胶囊乳液在室内91 d仍可检测到性信息素,而微胶囊乳液的野外持效期约为80 d。
应用昆虫性信息素防治害虫具有高效、安全无毒、不污染环境、不伤害有益生物等特点。昆虫性信息素配制成微胶囊能够增强有效物质的稳定性,通过改变微胶囊本身的各项系数(如囊壁厚度、粒径大小、壁材与芯材配制比例)达到缓慢释放性信息素的目的。目前,昆虫性信息素微胶囊技术在农林害虫绿色防控技术中扮演着日益重要的角色,配制成微胶囊后壁材不与性信息素产生化学作用,不会导致性信息素发生异构,能够确保性信息素不被外界环境所干扰[25-28],可产生非常可观的生态效益和社会效益,在农林业害虫防治中已经得到了一定的应用。例如:徐妍等[29]使用乙基纤维素作为囊壳,应用相分离法配制梨小食心虫性信息素微囊粒剂,比较了不同条件对微囊粒剂的平均粒径和包封率的影响,进行了室内缓释试验,试验结果显示,在室温时,微囊粒剂可以持续释放约110 d;李波等[30]分别使用微胶囊缓释剂、梨小食心虫硅橡胶性诱芯Y(北京中捷四方有限公司)和Isomate M Rosso(Pacific Biocontrol Corp, USA)进行迷向效果比较试验,微胶囊表现较好;利用微胶囊高空喷洒和喷雾喷洒,通过蜡滴将性信息素包埋并进行迷向试验,同样能够取得不错的效果[31]。除此之外,微胶囊包埋工艺技术在小菜蛾Plutella xylostella、松毛虫Dendrolimus及暗黑鳃金龟Holotrichia parallela性信息素制备等方面也有相关研究[32-34]。
在不同剪切速度和不同均质压力2个条件下,样品粒径分布图的双峰现象比较明显,出现双峰的原因可能是这2个因素对乳液均匀度起着关键性作用[35-36],低值剪切速度、压力的力度不够使乳液达到均匀,而过高的剪切速度和力度容易破坏乳液的稳定平衡。在不同的剪切速度和压力下,双峰的变化比较明显,说明剪切速度和均质压力对粒子分布集中度的影响比较大,当剪切速度变大时,粒径分布相对集中,而压力变大时粒径的分布却易分散,其可能原因是过高的压力破坏了乳液的稳定性,使乳液产生了部分聚集。
液体石蜡对微胶囊乳液粒径分布的影响表明:加入液体石蜡,可以在微胶囊外部形成一层保护膜,进而增加微胶囊壁厚度,减缓微胶囊芯材的释放速率。随着石蜡加入量的增多,粒径越来越小,包封率升高,乳液均匀稳定,但液体石蜡加入量过多时,易导致包封率下降[23]。因此,适当的液体石蜡含量可提高乳液的稳定性。
野外迷向试验中,80 d后4个处理的迷向率开始降低,主要原因是过了梨小食心虫发生的高峰期,新疆地区梨小食心虫在6月中旬达到第1代成虫羽化高峰,7月中旬为第 2 代成虫羽化高峰,而微胶囊乳液药效可以维持80 d左右,因此可有效防控梨小食心虫2代,实现跨时空覆盖,大大降低了梨小食心虫的危害;由于在77 d前,处理1(性信息素含量为45 g/hm2)和处理3(性信息素微胶囊乳液含量120 g/hm2)有迷向率较低的情况,因此综合考虑得出:处理2(性信息素含量为75 g/hm2)和处理4(性信息素微胶囊乳液含量75 g/hm2,冷藏1年)的迷向效果较佳,但4个处理的迷向效果无显著差异,说明冷藏条件不会对微胶囊乳液的野外迷向效果及持效时间造成太大影响,但微胶囊乳液经过冷藏,常常会发生聚合现象,致使乳液粒径和黏度变大,尤其是黏度较大会导致乳液流动性较差[37]。
梨小食心虫性信息素经微胶囊包埋技术处理后,延长了性信息素的作用持效期,性信息素微胶囊乳液可对梨小食心虫危害期进行时空全覆盖,持续降低虫口密度,引诱效果理想,对环境无污染,省时省力。因此,性信息素迷向法可以作为一种高效防控梨小食心虫的绿色防控技术,进而大面积应用示范推广。
-
-
[1] BRUCKNER R. Advanced organic chemistry: Reaction mechanisms [M]. Freiburg: Elsevier, 2001.
[2] BROWN W H, IVERSON B L, ANSLYN E, et al. Organic chemistry [M]. Boston: Cengage Learning, 2022.
[3] ZHAO L, WANG Y, ZHAO X, et al. Facile synthesis of nitrogen-doped carbon quantum dots with chitosan for fluorescent detection of Fe3+[J]. Polymers, 2019, 11(11): 1731. doi: 10.3390/polym11111731
[4] KATRITZKY A R, RAMSDEN C A,JOULE J A, et al. Handbook of heterocyclic chemistry[M]. New York: Elsevier, 2010.
[5] BASER K H. Biological and pharmacological activities of carvacrol and carvacrol bearing essential oils[J]. Current Pharmaceutical Design, 2008, 14(29): 3106-3119. doi: 10.2174/138161208786404227
[6] ORAV A, RAAL A, ARAK E. Essential oil composition of Pimpinella anisum L. fruits from various European countries[J]. Natural Product Research, 2008, 22(3): 227-232. doi: 10.1080/14786410701424667
[7] BECHER P G, VERSCHUT V, BIBB M J, et al. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal[J]. Nature Microbiology, 2020, 5(6): 821-829. doi: 10.1038/s41564-020-0697-x
[8] PARKER J K. Introduction to aroma compounds in foods[M]. New York: Elsevier, 2015: 3-30.
[9] COSTA D C, COSTA H S, ALBUQUERQUE T G, et al. Advances in phenolic compounds analysis of aromatic plants and their potential applications[J]. Trends in Food Science & Technology, 2015, 45(2): 336-354.
[10] BURT S. Essential oils: Their antibacterial properties and potential applications in foods: A review[J]. International Journal of Food Microbiology, 2004, 94: 223-253. doi: 10.1016/j.ijfoodmicro.2004.03.022
[11] SHI S, WANG Z, SHEN L, et al. Synthetic biology: A new frontier in food production[J]. Trends in Biotechnology, 2022, 40(7): 781-803. doi: 10.1016/j.tibtech.2022.01.002
[12] MCKAY D L, BLUMBERG J B. A review of the bioactivity and potential health benefits of chamomile tea (Matricaria recutita L. )[J]. Phytotherapy Research, 2006, 20(7): 519-530.
[13] HO S S M, KWONG A N L, WAN K W S, et al. Experiences of aromatherapy massage among adult female cancer patients: A qualitative study[J]. Journal of Clinical Nursing, 2017, 26(23/24): 4519-4526.
[14] KUMAR Y, PRAKASH O, TRIPATHI H, et al. AromaDb: A database of medicinal and aromatic plant's aroma molecules with phytochemistry and therapeutic potentials[J]. Frontiers in Plant Science, 2018, 9: 1081. doi: 10.3389/fpls.2018.01081
[15] CERONI F, ELLIS T. The challenges facing synthetic biology in eukaryotes[J]. Nature Reviews Molecular Cell Biology, 2018, 19(8): 481-482. doi: 10.1038/s41580-018-0013-2
[16] NIELSEN J, KEASLING J D. Engineering cellular metabolism[J]. Cell, 2016, 164(6): 1185-1197. doi: 10.1016/j.cell.2016.02.004
[17] HUGHES R A, ELLINGTON A D. Synthetic DNA synthesis and assembly: Putting the synthetic in synthetic biology[J]. Cold Spring Harbor Perspectives in Biology, 2017, 9(1): a023812. doi: 10.1101/cshperspect.a023812
[18] KE J, WANG B, YOSHIKUNI Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture[J]. Trends in Biotechnology, 2021, 39(3): 244-261. doi: 10.1016/j.tibtech.2020.07.008
[19] DOU J, BENNETT M R. Synthetic biology and the gut microbiome[J]. Biotechnology Journal, 2018, 13(5): 1700159. doi: 10.1002/biot.201700159
[20] SMANSKI M J, ZHOU H, CLAESEN J, et al. Synthetic biology to access and expand nature’s chemical diversity[J]. Nature Reviews Microbiology, 2016, 14(3): 135-149. doi: 10.1038/nrmicro.2015.24
[21] THODEY K, GALANIE S, SMOLKE C D. A microbial biomanufacturing platform for natural and semisynthetic opioids[J]. Nature Chemical Biology, 2014, 10(10): 837-844. doi: 10.1038/nchembio.1613
[22] TAN C, XU P, TAO F. Carbon-negative synthetic biology: Challenges and emerging trends of cyanobacterial technology[J]. Trends in Biotechnology, 2022, 40(12): 1488-1502. doi: 10.1016/j.tibtech.2022.09.012
[23] MORROW G W. The shikimate pathway: Biosynthesis of phenolic products from shikimic acid[M]. New York: Oxford University Press, 2016.
[24] JIANG M, ZHANG H. Engineering the shikimate pathway for biosynthesis of molecules with pharmaceutical activities in E. coli[J]. Current Opinion in Biotechnology, 2016, 42: 1-6. doi: 10.1016/j.copbio.2016.01.016
[25] STARCEVIC A, AKTHAR S, DUNLAP W C, et al. Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(7): 2533-2537.
[26] MAEDA H, DUDAREVA N. The shikimate pathway and aromatic amino acid biosynthesis in plants[J]. Annual Review of Plant Biology, 2012, 63: 73-105. doi: 10.1146/annurev-arplant-042811-105439
[27] PITTARD J, YANG J. Biosynthesis of the aromatic amino acids[J]. EcoSal Plus, 2008, 3(1): 1110-1128.
[28] YOKOYAMA R, KLEVEN B, GUPTA A, et al. 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase as the gatekeeper of plant aromatic natural product biosynthesis[J]. Current Opinion in Plant Biology, 2022, 67: 102219. doi: 10.1016/j.pbi.2022.102219
[29] 向莉, 李盾. 达菲的主要合成中间体莽草酸获得的新进展[J]. 医药产业资讯, 2006, 6: 52-42. [30] JANSEN F, GILLESSEN B, MUELLER F, et al. Metabolic engineering for p-coumaryl alcohol production in Escherichia coli by introducing an artificial phenylpropanoid pathway[J]. Biotechnology and Applied Biochemistry, 2014, 61(6): 646-654. doi: 10.1002/bab.1222
[31] 鄢芳清, 韩亚昆, 李娟, 等. 大肠杆菌芳香族氨基酸代谢工程研究进展[J]. 生物加工过程, 2017, 15(5): 32-39. [32] ADAMS Z P, EHLTING J, EDWARDS R. The regulatory role of shikimate in plant phenylalanine metabolism[J]. Journal of Theoretical Biology, 2019, 462: 158-170. doi: 10.1016/j.jtbi.2018.11.005
[33] LUTTIK M A H, VURALHAN Z, SUIR E, et al. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: Quantification of metabolic impact[J]. Metabolic Engineering, 2008, 10(3): 141-53.
[34] TZIN V, MALITSKY S, ZVI M M B, et al. Expression of a bacterial feedback-insensitive 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase of the shikimate pathway in Arabidopsis elucidates potential metabolic bottlenecks between primary and secondary metabolism[J]. New Phytologist, 2012, 194(2): 430-439. doi: 10.1111/j.1469-8137.2012.04052.x
[35] YOKOYAMA R, DE OLIVEIRA M V V, KLEVEN B, et al. The entry reaction of the plant shikimate pathway is subjected to highly complex metabolite-mediated regulation[J]. Plant Cell, 2021, 33(3): 671-696. doi: 10.1093/plcell/koaa042
[36] HU C, JIANG P, XU J, et al. Mutation analysis of the feedback inhibition site of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase of Escherichia coli[J]. Journal of Basic Microbiology, 2003, 43(5): 399-406.
[37] YOKOYAMA R, DE OLIVEIRA M V V, TAKEDA-KIMURA Y, et al. Point mutations that boost aromatic amino acid production and CO2 assimilation in plants[J]. Science Advances, 2022, 8(23): eabo3416. doi: 10.1126/sciadv.abo3416
[38] VOGT T. Phenylpropanoid biosynthesis[J]. Molecular Plant, 2010, 3(1): 2-20. doi: 10.1093/mp/ssp106
[39] BORTESI L, FISCHER R. The CRISPR/Cas9 system for plant genome editing and beyond[J]. Biotechnology Advances, 2015, 33(1): 41-52. doi: 10.1016/j.biotechadv.2014.12.006
[40] POTT D M, OSORIO S, VALLARINO J G. From central to specialized metabolism: An overview of some secondary compounds derived from the primary metabolism for their role in conferring nutritional and organoleptic characteristics to fruit[J]. Frontiers in Plant Science, 2019, 10: 835. doi: 10.3389/fpls.2019.00835
[41] YANG D, DU X, YANG Z, et al. Transcriptomics, proteomics, and metabolomics to reveal mechanisms underlying plant secondary metabolism[J]. Engineering in Life Sciences, 2014, 14(5): 456-466. doi: 10.1002/elsc.201300075
[42] FU R, MARTIN C, ZHANG Y. Next-generation plant metabolic engineering, inspired by an ancient Chinese irrigation system[J]. Molecular Plant, 2018, 11(1): 47-57. doi: 10.1016/j.molp.2017.09.002
[43] ASHOKKUMAR S, JAGANATHAN D, RAMANATHAN V, et al. Creation of novel alleles of fragrance gene OsBADH2 in rice through CRISPR/Cas9 mediated gene editing[J]. PLoS One, 2020, 15(8): e0237018. doi: 10.1371/journal.pone.0237018
[44] HOFFMANN T, KURTZER R, SKOWRANEK K, et al. Metabolic engineering in strawberry fruit uncovers a dormant biosynthetic pathway[J]. Metabolic Engineering, 2011, 13(5): 527-531. doi: 10.1016/j.ymben.2011.06.002
[45] LOBATO-GÓMEZ M, HEWITT S, CAPELL T, et al. Transgenic and genome-edited fruits: Background, constraints, benefits, and commercial opportunities[J]. Horticulture Research, 2021, 8(1): 166.
[46] YU J, TU L, SUBBURAJ S, et al. Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins[J]. Plant Cell Reports, 2021, 40: 1037-1045. doi: 10.1007/s00299-020-02593-1
[47] AKHTAR T A, PICHERSKY E. Veratrole biosynthesis in white campion[J]. Plant Physiology, 2013, 162(1): 52-62. doi: 10.1104/pp.113.214346
[48] FARHI M, LAVIE O, MASCI T, et al. Identification of rose phenylacetaldehyde synthase by functional complementation in yeast[J]. Plant Molecular Biology, 2010, 72: 235-245. doi: 10.1007/s11103-009-9564-0
[49] LIAO P, RAY S, BOACHON B, et al. Cuticle thickness affects dynamics of volatile emission from petunia flowers[J]. Nature Chemical Biology, 2021, 17(2): 138-145. doi: 10.1038/s41589-020-00670-w
[50] SPITZER-RIMON B, FARHI M, ALBO B, et al. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia[J]. Plant Cell, 2012, 24(12): 5089-5105.
[51] GURURAJ H B, PADMA M N, GIRIDHAR P, et al. Functional validation of Capsicum frutescens aminotransferase gene involved in vanillylamine biosynthesis using Agrobacterium mediated genetic transformation studies in Nicotiana tabacum and Capsicum frutescens calli cultures[J]. Plant Science, 2012, 195: 96-105. doi: 10.1016/j.plantsci.2012.06.014
[52] WEBER N, ISMAIL A, GORWA-GRAUSLUND M, et al. Biocatalytic potential of vanillin aminotransferase from Capsicum chinense[J]. BMC Biotechnology, 2014, 14: 1-6.
[53] GALLAGE N J, HANSEN E H, KANNANGARA R, et al. Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme[J]. Nature Communications, 2014, 5: 4037.
[54] GASSON M J, KITAMURA Y, MCLAUCHLAN W R, et al. Metabolism of ferulic acid to vanillin: A bacterial gene of the enoyl-SCoA hydratase/isomerase superfamily encodes an enzyme for the hydration and cleavage of a hydroxycinnamic acid SCoA thioester[J]. Journal of Biological Chemistry, 1998, 273(7): 4163-4170. doi: 10.1074/jbc.273.7.4163
[55] SINGH P, KHAN S I, PANDEY S S, et al. Vanillin production in metabolically engineered Beta vulgaris hairy roots through heterologous expression of Pseudomonas fluorescens HCHL gene[J]. Industrial Crops and Products, 2015, 74: 839-848. doi: 10.1016/j.indcrop.2015.05.037
[56] KUNDU A. Vanillin biosynthetic pathways in plants[J]. Planta, 2017, 245(6): 1069-1078. doi: 10.1007/s00425-017-2684-x
[57] MAYER M J, NARBAD A, PARR A J, et al. Rerouting the plant phenylpropanoid pathway by expression of a novel bacterial enoyl-CoA hydratase/lyase enzyme function[J]. Plant Cell, 2001, 13(7): 1669-1682. doi: 10.1105/TPC.010063
[58] KNUDSEN J, ERIKSSON R, GERSHENZON J, et al. Diversity and distribution of floral scent[J]. Botanical Review, 2006, 72: 1-120. doi: 10.1663/0006-8101(2006)72[1:DADOFS]2.0.CO;2
[59] OLIVA M, BAR E, OVADIA R, et al. Phenylpyruvate contributes to the synthesis of fragrant benzenoid-phenylpropanoids in Petunia× hybrida flowers[J]. Frontiers in Plant Science, 2017, 8: 769. doi: 10.3389/fpls.2017.00769
[60] SKALITER O, RAVID J, SHKLARMAN E, et al. Ectopic expression of PAP1 leads to anthocyanin accumulation and novel floral color in genetically engineered goldenrod (Solidago canadensis L.)[J]. Frontiers in Plant Science, 2019, 10: 1561. doi: 10.3389/fpls.2019.01561
[61] CNA’ ANI A, MÜHLEMANN J K, RAVID J, et al. Petunia × hybrida floral scent production is negatively affected by high-temperature growth conditions[J]. Plant, Cell & Environment, 2015, 38(7): 1333-1346.
[62] KOEDUKA T, TAKARADA S, FUJII K, et al. Production of raspberry ketone by redirecting the metabolic flux to the phenylpropanoid pathway in tobacco plants[J]. Metabolic Engineering Communications, 2021, 13: e00180. doi: 10.1016/j.mec.2021.e00180
[63] YOSHIDA K, OYAMA-OKUBO N, YAMAGISHI M. An R2R3-MYB transcription factor ODORANT1 regulates fragrance biosynthesis in lilies (Lilium spp. )[J]. Molecular Breeding, 2018, 38(144): 1-14.
[64] BOERSMA M R, PATRICK R M, JILLINGS S L, et al. ODORANT1 targets multiple metabolic networks in petunia flowers[J]. The Plant Journal, 2022, 109(5): 1134-1151. doi: 10.1111/tpj.15618
[65] SHOR E, RAVID J, SHARON E, et al. SCARECROW-like GRAS protein PES positively regulates petunia floral scent production[J]. Plant Physiology, 2023, 192(1): 409-425. doi: 10.1093/plphys/kiad081
[66] TZIN V, ROGACHEV I, MEIR S, et al. Altered levels of aroma and volatiles by metabolic engineering of shikimate pathway genes in tomato fruits[J]. AIMS Bioengineering, 2015, 2(2): 75-92. doi: 10.3934/bioeng.2015.2.75
[67] XIE Q, LIU Z, MEIR S, et al. Altered metabolite accumulation in tomato fruits by coexpressing a feedback-insensitive AroG and the PhODO1 MYB-type transcription factor[J]. Plant Biotechnology Journal, 2016, 14(12): 2300-2309. doi: 10.1111/pbi.12583
[68] PATTERSON E L, PETTINGA D J, RAVET K, et al. Glyphosate resistance and EPSPS gene duplication: Convergent evolution in multiple plant species[J]. Journal of Heredity, 2018, 109(2): 117-125. doi: 10.1093/jhered/esx087
[69] CHHAPEKAR S, RAGHAVENDRARAO S, PAVAN G, et al. Transgenic rice expressing a codon-modified synthetic CP4-EPSPS confers tolerance to broad-spectrum herbicide, glyphosate[J]. Plant Cell Reports, 2015, 34: 721-731. doi: 10.1007/s00299-014-1732-2
[70] 李国平, 刘 冰, 黄建荣, 等. 转聚合cry1A.105、cry2Ab2和cp4epsps基因抗虫耐除草剂玉米的田间抗性评价[J]. 植物保护, 2019, 45(1): 142-147. [71] LIANG C, SUN B, MENG Z, et al. Co-expression of GR79 EPSPS and GAT yields herbicide-resistant cotton with low glyphosate residues[J]. Plant Biotechnology Journal, 2017, 15(12): 1622-1629. doi: 10.1111/pbi.12744
[72] YANNICCARI M, VÁZQUEZ-GARCÍA J G, GIGÓN R, et al. A novel EPSPS Pro-106-His mutation confers the first case of glyphosate resistance in Digitaria sanguinalis[J]. Pest Management Science, 2022, 78(7): 3135-3143. doi: 10.1002/ps.6940
[73] ENDO M, MIKAMI M, ENDO A, et al. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM[J]. Nature Plants, 2019, 5(1): 14-17.
[74] LI H, LI J, CHEN J, et al. Precise modifications of both exogenous and endogenous genes in rice by prime editing[J]. Molecular Plant, 2020, 13(5): 671-674. doi: 10.1016/j.molp.2020.03.011
[75] 林春草, 陈大伟, 戴均贵. 黄酮类化合物合成生物学研究进展[J]. 药学学报, 2022, 57(5): 1322-1335. [76] 沈忠伟, 许昱, 夏犇, 等. 植物类黄酮次生代谢生物合成相关转录因子及其在基因工程中的应用 [J]. 2008, 6(3): 542-548. [77] DENG X, BASHANDY H, AINASOJA M, et al. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida[J]. New Phytologist, 2014, 201(4): 1469-1483. doi: 10.1111/nph.12610
[78] ZHANG X, ABRAHAN C, COLQUHOUN T A, et al. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis[J]. Plant Cell, 2017, 29(5): 1157-1174. doi: 10.1105/tpc.16.00855
[79] SCHIJLEN E G, DE VOS C H, MARTENS S, et al. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits[J]. Plant Physiology, 2007, 144(3): 1520-1530. doi: 10.1104/pp.107.100305
[80] BOMATI E K, AUSTIN M B, BOWMAN M E, et al. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis[J]. Journal of Biological Chemistry, 2005, 280(34): 30496-30503. doi: 10.1074/jbc.M502239200
[81] SHIMADA N, NAKATSUKA T, NISHIHARA M, et al. Isolation and characterization of a cDNA encoding polyketide reductase in Lotus japonicus[J]. Plant Biotechnology, 2006, 23(5): 509-513. doi: 10.5511/plantbiotechnology.23.509
[82] YIN Y, ZHANG X, GAO Z, et al. The research progress of chalcone isomerase (CHI) in plants[J]. Molecular Biotechnology, 2019, 61: 32-52. doi: 10.1007/s12033-018-0130-3
[83] ZHU J, ZHAO W, LI R, et al. Identification and characterization of chalcone isomerase genes involved in flavonoid production in Dracaena cambodiana[J]. Frontiers in Plant Science, 2021, 12: 616396. doi: 10.3389/fpls.2021.616396
[84] WANG H, LIU S, WANG T, et al. The moss flavone synthase I positively regulates the tolerance of plants to drought stress and UV-B radiation[J]. Plant Science, 2020, 298: 110591. doi: 10.1016/j.plantsci.2020.110591
[85] LIU W, FENG Y, YU S, et al. The flavonoid biosynthesis network in plants[J]. International Journal of Molecular Sciences, 2021, 22: 12824. doi: 10.3390/ijms222312824
[86] YE J H, LV Y Q, LIU S R, et al. Effects of light intensity and spectral composition on the transcriptome profiles of leaves in shade grown tea plants (Camellia sinensis L. ) and regulatory network of flavonoid biosynthesis[J]. Molecules, 2021, 26(19): 5836. doi: 10.3390/molecules26195836
[87] XU W, DUBOS C, LEPINIEC L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends in Plant Science, 2015, 20(3): 176-185. doi: 10.1016/j.tplants.2014.12.001
[88] LI C, QIU J, DING L, et al. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals[J]. Plant Physiology and Biochemistry, 2017, 112: 335-345. doi: 10.1016/j.plaphy.2017.01.019
[89] BOVY A, DE VOS R, KEMPER M, et al. High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1[J]. The Plant Cell, 2002, 14(10): 2509-2526. doi: 10.1105/tpc.004218
[90] GAO Y, LIU J, CHEN Y, et al. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins[J]. Horticulture Research, 2018, 5: 11-18. doi: 10.1038/s41438-018-0016-3
[91] WANG J, LI G, LI C, et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato[J]. New Phytologist, 2021, 229(6): 3237-3252. doi: 10.1111/nph.17112
[92] VANHOLME R, DEMEDTS B, MORREEL K, et al. Lignin biosynthesis and structure[J]. Plant Physiology, 2010, 153(3): 895-905. doi: 10.1104/pp.110.155119
[93] VANHOLME B, DESMET T, RONSSE F, et al. Towards a carbon-negative sustainable bio-based economy[J]. Frontiers in Plant Science, 2013, 4: 174.
[94] SCHUTYSER W, RENDERS T, VAN DEN BOSCH S, et al. Chemicals from lignin: An interplay of lignocellulose fractionation, depolymerisation, and upgrading[J]. Chemical Society Reviews, 2018, 47(3): 852-908. doi: 10.1039/C7CS00566K
[95] DE MEESTER B, VANHOLME R, MOTA T, et al. Lignin engineering in forest trees: From gene discovery to field trials[J]. Plant Communications, 2022, 3(6): 100465. doi: 10.1016/j.xplc.2022.100465
[96] CHANOCA A, DE VRIES L, BOERJAN W. Lignin engineering in forest trees[J]. Frontiers in Plant Science, 2019, 10: 912. doi: 10.3389/fpls.2019.00912
[97] WANG H, XUE Y, CHEN Y, et al. Lignin modification improves the biofuel production potential in transgenic Populus tomentosa[J]. Industrial Crops and Products, 2012, 37(1): 170-177. doi: 10.1016/j.indcrop.2011.12.014
[98] STOUT A T, DAVIS A A, DOMEC J C, et al. Growth under field conditions affects lignin content and productivity in transgenic Populus trichocarpa with altered lignin biosynthesis[J]. Biomass and Bioenergy, 2014, 68: 228-239. doi: 10.1016/j.biombioe.2014.06.008
[99] VAN ACKER R, DEJARDIN A, DESMET S, et al. Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1[J]. Plant Physiology, 2017, 175(3): 1018-1039. doi: 10.1104/pp.17.00834
[100] XIANG Z, SEN S K, MIN D, et al. Field-grown transgenic hybrid poplar with modified lignin biosynthesis to improve enzymatic saccharification efficiency[J]. ACS Sustainable Chemistry & Engineering, 2017, 5(3): 2407-2414.
[101] SALEME M D S, CESARINO I, VARGAS L, et al. Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar[J]. Plant Physiology, 2017, 175(3): 1040-1057. doi: 10.1104/pp.17.00920
[102] DE VRIES L, BROUCKAERT M, CHANOCA A, et al. CRISPR-Cas9 editing of CAFFEOYL SHIKIMATE ESTERASE 1 and 2 shows their importance and partial redundancy in lignification in Populus tremula × P. alba[J]. Plant Biotechnology Journal, 2021, 19(11): 2221-2234. doi: 10.1111/pbi.13651
[103] DE MEESTER B, MADARIAGA CALDERÓN B, DE VRIES L, et al. Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele[J]. Nature Communications, 2020, 11(1): 5020. doi: 10.1038/s41467-020-18822-w
[104] CAO S, HUANG C, LUO L, et al. Cell-specific suppression of 4-coumarate-CoA ligase gene reveals differential effect of lignin on cell physiological function in Populus[J]. Frontiers in Plant Science, 2020, 11: 589729. doi: 10.3389/fpls.2020.589729
[105] GUI J S, LAM P Y, TOBIMATSU Y, et al. Fibre-specific regulation of lignin biosynthesis improves biomass quality in Populus[J]. New Phytologist, 2020, 226(4): 1074-1087. doi: 10.1111/nph.16411
[106] SULIS D B, JIANG X, YANG C, et al. Multiplex CRISPR editing of wood for sustainable fiber production[J]. Science, 2023, 381(6654): 216-221. doi: 10.1126/science.add4514
[107] STEPANYUK A, KIRSCHNING A. Synthetic terpenoids in the world of fragrances: Iso E Super® is the showcase[J]. Beilstein Journal of Organic Chemistry, 2019, 15: 2590-2602.
-
期刊类型引用(2)
1. 温强,程晓琴,李晓雪,王珊珊,姚志超,张宏宇. 实蝇觅食、交配和产卵行为相关的信息化合物研究及其应用进展. 应用昆虫学报. 2023(02): 426-448 .  百度学术
百度学术
2. 相会明,刁红亮,李先伟,赵志国,马瑞燕. 梨小食心虫性信息素在四种载体中释放速率及田间迷向率研究. 应用昆虫学报. 2022(02): 311-317 .  百度学术
百度学术
其他类型引用(5)



 下载:
下载: