Morphological characterization and genetic mapping of shrunken endosperm mutant sh2021 in maize
-
摘要:目的
分析玉米籽粒皱缩突变体的表型特征并进行籽粒相关基因的精细定位,为揭示该基因调控玉米籽粒发育的分子机制奠定基础。
方法以玉米自交系‘B73’种植过程中籽粒自发突变个体为材料,命名为shank2021(sh2021),对其形态学和细胞学特征进行观察;构建分离群体,通过混合群体分离分析法(Bulked segregant analysis,BSA)对基因进行初步定位,筛选交换单株进一步缩小定位区间,最后结合转录组测序及基因功能注释推测控制籽粒缺陷性状的候选基因。
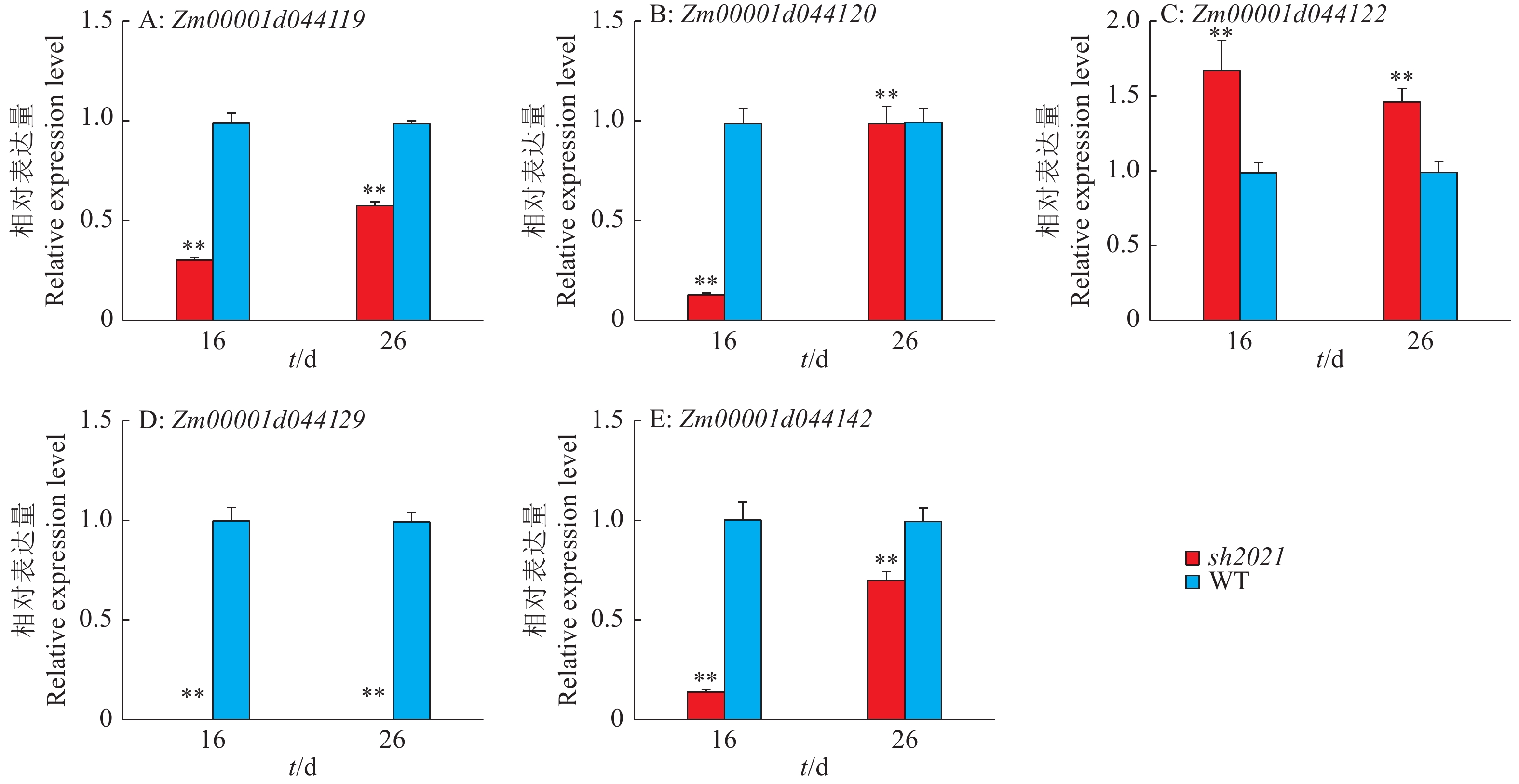
结果与野生型相比,sh2021籽粒凹陷皱缩、颜色加深、籽粒排列不规则,且百粒质量降低。扫描电镜观察发现,与野生型相比,sh2021胚乳细胞和淀粉粒均显著变小,且淀粉粒大小不均匀。遗传分析结果表明,sh2021是由单个隐性基因突变所致。利用BSA分析方法将目的基因定位在3号染色体末端约13.25 Mb区域。进一步扩大分离群体筛选交换单株,将目的基因定位在标记ID5与ID9之间的529.60 kb范围。通过sh2021和野生型的转录组测序,结合基因功能注释结果,预测Zm00001d044119、Zm00001d044120、Zm00001d044122、Zm00001d044129、Zm00001d044142这5个基因可能是影响玉米籽粒发育的重要候选基因。
结论sh2021的发现与鉴定为进一步深入解析玉米籽粒发育的遗传机制提供了丰富的试验材料,同时也为后续相关基因的克隆和功能解析奠定了基础。
Abstract:ObjectiveTo analyze the phenotypic characteristic of maize shrunk endosperm mutant and fine mapping of related genes, and lay a foundation for further understanding molecular mechanism of maize kernel development.
MethodThe spontaneous shrunk endosperm mutant shank2021(sh2021) was isolated from maize inbred line B73. Morphological and cytological characteristics were observed. A segregating population was developed, and the bulked segregant analysis (BSA) was used for preliminary gene mapping. The recombinant plants were screened for further narrowing the mapping interval. Finally the candidate genes controlling grain defect traits were speculated by transcriptome sequencing combined with gene function annotation analysis.
ResultCompared with wild type, sh2021 displayed sunken and shrunken kernels, darker color, irregular grain arrangement, and lower 100-grain weight. The scanning electron microscope observation revealed that sh2021 had smaller endosperm cells and starch granules, and the starch granules were varied in size. The genetic analysis results indicated that sh2021 was caused by a single recessive gene mutation. The BSA indicated that the target gene was located on a 13.25 Mb fragment at the end of chromosome 3. By further expanding the segregating population and screening recombinant plants, the target gene was narrowed down to an interval of 529.60 kb between markers ID5 and ID9. Transcriptome sequencing and gene annotation of sh2021 and wild type indicated that Zm00001d044119, Zm00001d044120, Zm00001d044122, Zm00001d044129, and Zm00001d044142 mighted be candidate genes controlling maize kernel development.
ConclusionThe identification of sh2021 provides abundant experimental materials for the study of kernel development, and lays a foundation for further map-based cloning and functional analysis of sh2021.
-
Keywords:
- Maize /
- Shrunken endosperm mutant /
- Genetic analysis /
- Fine mapping
-
-
图 1 sh2021突变体表型鉴定(F2分离群体)
A:完熟期玉米果穗;B:籽粒表型;C:籽粒百粒质量;“**”表示sh201与野生型在0.01水平差异显著(t检验)
Figure 1. Phenotypic identification of sh2021 mutants (F2 segregating population)
A: Mature ear; B: Grain phenotype; C: 100-kernel weight; “**” indicate significant difference between wild type and sh2021 at the 0.01 level (t test)
图 4 SNP-index的全基因组分布图
散点为原始值,黑色曲线为拟合值;蓝色阈值线和红色阈值线分别表示各位点以及窗口的ΔSNP-index 95%和99%置信区间;横坐标为染色体的分布,每个点代表ΔSNP-index值
Figure 4. Genome-wide distribution of SNP-index
The scatter points are the original values, and the black curve is the fitted value; Blue threshold line and red threshold line indicate the ΔSNP-index 95% and 99% confidence intervals, respectively; The horizontal coordinate is the distribution of chromosomes, and each dot represents the ΔSNP-index value
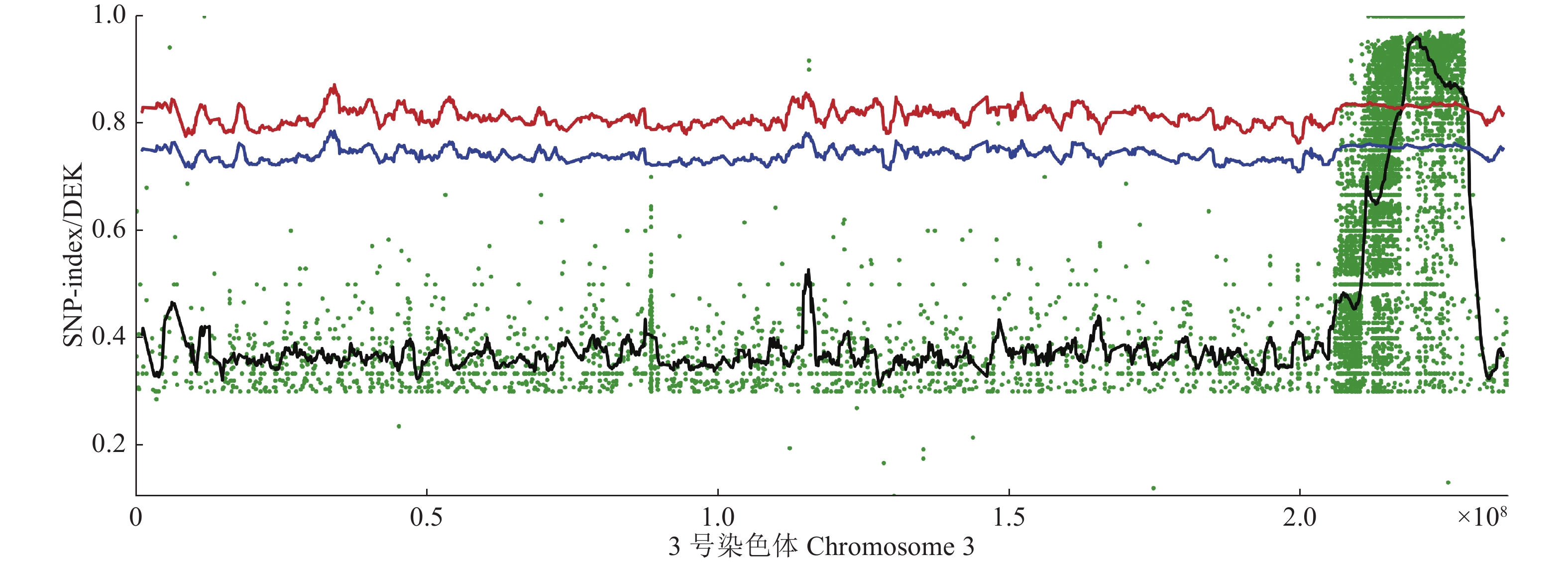
图 5 SNP-index在3号染色体上的分布
散点图为原始值,黑色曲线为拟合值,其他横线为指定阈值位置;横坐标为3号染色体的分布,每个点代表的ΔSNP-index值
Figure 5. Distribution of SNP-index on Chromosome 3
The scatter plot is the original value, the black curve is the fitted value, and the other horizontal lines are the specified threshold positions; The horizontal coordinate is the distribution of Chr 3, and each dot represents the ΔSNP-index value
表 1 sh2021 基因定位引物
Table 1 Primers used for gene mapping of sh2021
引物名称
Primer name序列(5′→3′)
SequenceID1 F:TACAGCTTGTAAAAATACAGGGCC
R:ATGAGAGTGCATGGTCCGTGID2 F:GGCAGCAGGATCAGAAGAGA
R:GGAACTTGTTGTGCCAAGGID3 F:TCTCGAATCAAGAACCAGCA
R:GGAGGGATTGGGTGAAGATTID4 F:GTCTACAACCGGCTCCTCAA
R;GTGGAAGTGTCGTCCGTTCTID5 F:GCCATCTACAGTGTCAGCCA
R:GTCAACGAGATTCTCAGGGCID9 F:CTTCCCGTCGAGATCCACG
R:GTTGACAGCTTGTGGGGTCTID10 F:GCTAGAGATGGCTATTAGGTGCA
R:AGCGTGTCTAACCTCACTTATAACAID11 F:TAACTGTCTCGCTGGGCTTT
R:GGCGTCCGTCCAAATAAAID12 F:CCATGAGGAGGCATGTGTGG
R:CAGCTGGTTCACGCCACATCID13 F:TGCACTTCGAGAGGTTTGTT
R:TCATAATAGTCTCGTATCCAGCCTID14 F:ACCCAAGGACCCCAAAGAGA
R:GCCCTATGCTTGGGGAGTGID15 F:GCCTGTCTAGCATATGATGTGAA
R:GTCTCATGCCAAACGAGGATC3-102 F:CTAGCCAGCCCCCATTCTTC
R:GCAAGGAGTAGGGAGGACGTG表 2 qRT-PCR定量验证引物
Table 2 qRT-PCR quantitative verification primers
引物名称
Primer name序列(5′→3′)
SequenceHX1 F:TTTGAGCGAAACTCCAGTGC
R:ATGTTCATGTGCTGCAGGACHX2 F:GAGTCGGAGATCAGCATCGG
R:AAAGTGTCGGCGAAGAACCTHX3 F:TTGGGAAGACGAAGCCATTG
R:AGGTCGGCTTTCCATATGGACHX4 F:TCACTGAGCAGCCTTCCAAG
R:GACACGTGAGCAGACTCCAAHX5 F:GCCACGGGATCATGAAGGAG
R:TCTCCTCCAACAGACCCTCC -
[1] NEUFFER M G, SHERIDAN W F. Defective kernel mutants of maize: I: Genetic and lethality studies[J]. Genetics, 1980, 95(4): 929-944. doi: 10.1093/genetics/95.4.929
[2] BRUNELLE D C, CLARK J K, SHERIDAN W F. Genetic screening for EMS-induced maize embryo-specific mutants altered in embryo morphogenesis[J]. G3-Genes Genomics Genetics, 2017, 7(11): 3559-3570.
[3] WU Y R, MESSING J. Proteome balancing of the maize seed for higher nutritional value[J]. Frontiers in Plant Science, 2014, 5: 240.
[4] SCHMIDT R J, BURR F A, AUKERMAN M J, et al. Maize regulatory gene opaque-2 encodes a protein with a “leucine-zipper” motif that binds to zein DNA[J]. Proceedings of the National Academy of Sciences of the United States of America, 1990, 87(1): 46-50. doi: 10.1073/pnas.87.1.46
[5] AZEVEDO R A, LEA P J, DAMERVAL C, et al. Regulation of lysine metabolism and endosperm protein synthesis by the opaque-5 and opaque-7 maize mutations[J]. Journal of Agricultural and Food Chemistry, 2004, 52(15): 4865-4871. doi: 10.1021/jf035422h
[6] MYERS A M, JAMES M G, LIN Q. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function[J]. Plant Cell, 2011, 23(6): 2331-2347. doi: 10.1105/tpc.111.087205
[7] FU S N, SCANLON M J. Clonal mosaic analysis of empty pericarp2 reveals nonredundant functions of the duplicated heat shock factor binding proteins during maize shoot development[J]. Genetics, 2004, 167(3): 1381-1394. doi: 10.1534/genetics.104.026575
[8] CHETTOOR A M, YI G, GOMEZ E, et al. A putative plant organelle RNA recognition protein gene is essential for maize kernel development[J]. Journal of Integrative Plant Biology, 2015, 57(3): 236-246. doi: 10.1111/jipb.12234
[9] YANG Y Z, DING S, WANG Y, et al. Small kernel2 encodes a glutaminase in vitamin B6 biosynthesis essential for maize seed development[J]. Plant Physiology, 2017, 174(2): 1127-1138. doi: 10.1104/pp.16.01295
[10] 薛慧, 张国治, 吕飞杰, 等. 抗性淀粉测定方法的研究[J]. 河南工业大学学报(自然科学版), 2012, 33(4): 57-60. doi: 10.16433/j.cnki.issn1673-2383.2012.04.017 [11] DICKINSON D B, PREISS J. Presence of ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize endosperm[J]. Plant Physiology, 1969, 44(7): 1058-1062. doi: 10.1104/pp.44.7.1058
[12] 周瑞颐, 青芸, 李道杨, 等. 玉米ZmBT1研究进展[J]. 分子植物育种, 2020, 18(20): 6702-6706. doi: 10.13271/j.mpb.018.006702 [13] MAGALIE C, PIERRE C, SYLVIE M, et al. Transcriptional and metabolic adjustments in ADP-glucose pyrophosphorylase-deficient bt2 maize kernels[J]. Plant Physiology, 2008, 146(4): 1553-1570. doi: 10.1104/pp.107.112698
[14] 宋欣冉, 胡书婷, 张凯, 等. 玉米籽粒突变体dek101的表型分析和精细定位[J]. 作物学报, 2020, 46(12): 1831-1838. [15] 石慧敏, 蒋成功, 王红武, 等. 玉米籽粒突变体dek48的表型鉴定与基因定位[J]. 作物学报, 2020, 46(9): 1359-1367. [16] HE Y H, WANG J G, QI W W, et al. Maize Dek15 encodes the cohesin-loading complex subunit SCC4 and is essential for chromosome segregation and kernel development[J]. The Plant Cell, 2019, 31(2): 465-485. doi: 10.1105/tpc.18.00921
[17] WANG G F, WANG F, WANG G, et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm[J]. The Plant Cell, 2012, 24(8): 3447-3462. doi: 10.1105/tpc.112.101360
[18] WANG G, QI W W, WU Q, et al. Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly[J]. Plant Physiology, 2014, 165(2): 582-594. doi: 10.1104/pp.114.238030
[19] GILLIKIN J W, ZHANG F, COLEMAN C E, et al. A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane[J]. Plant Physiology, 1997, 114(1): 345-352. doi: 10.1104/pp.114.1.345
[20] KIM C S, GIBBON B C, GILLIKIN J W, et al. The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response1[J]. The Plant Journal, 2006, 48(3): 440-451. doi: 10.1111/j.1365-313X.2006.02884.x
[21] KUSHWAHA H R, SINGH A K, SOPORY S K, et al. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L. ) Heynh and Oryza sativa L. reveals their developmental and stress regulation[J]. BMC Genomics, 2009, 10: 200. doi: 10.1186/1471-2164-10-200
[22] 王立成. 玉米抗旱高通量FOX文库的构建与验证[D]. 杨凌: 西北农林科技大学, 2019. [23] 崔会婷, 蒋旭, 张铁军, 等. 植物CYP450家族研究进展[J]. 中国草地学报, 2020, 42(5): 173-180. doi: 10.16742/j.zgcdxb.20190182 [24] 刘薇, 张彦威, 李伟, 等. 大豆细胞色素P450基因GmCYP78A69的克隆和生物信息学分析[J]. 分子植物育种, 2020, 18(14): 4523-4531. [25] CHEN X, LIU W, HUANG X, et al. Arg-type dihydroflavonol 4-reductase genes from the fern Dryopteris erythrosora play important roles in the biosynthesis of anthocyanins[J]. PLoS One, 2020, 15(5): e232090. doi: 10.1371/journal.pone.0232090.
[26] GU Z Y, CHEN H, YANG R N, et al. Identification of DFR as a promoter of anthocyanin accumulation in poinsettia (Euphorbia pulcherrima, Willd. ex Klotzsch) bracts under short-day conditions[J]. Scientia Horticulturae, 2018, 236: 158-165. doi: 10.1016/j.scienta.2018.03.032
[27] DEIKMAN J, HAMMER P E. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana[J]. Plant Physiology, 1995, 108(1): 47-57. doi: 10.1104/pp.108.1.47
[28] BAI M Y, FAN M, OH E, et al. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis[J]. The Plant Cell, 2012, 24(12): 4917-4929.
[29] 刘浩. 转录因子ABP7在玉米籽粒发育过程中的功能及其分子机理分析[D]. 北京: 中国农业大学, 2017.




 下载:
下载: