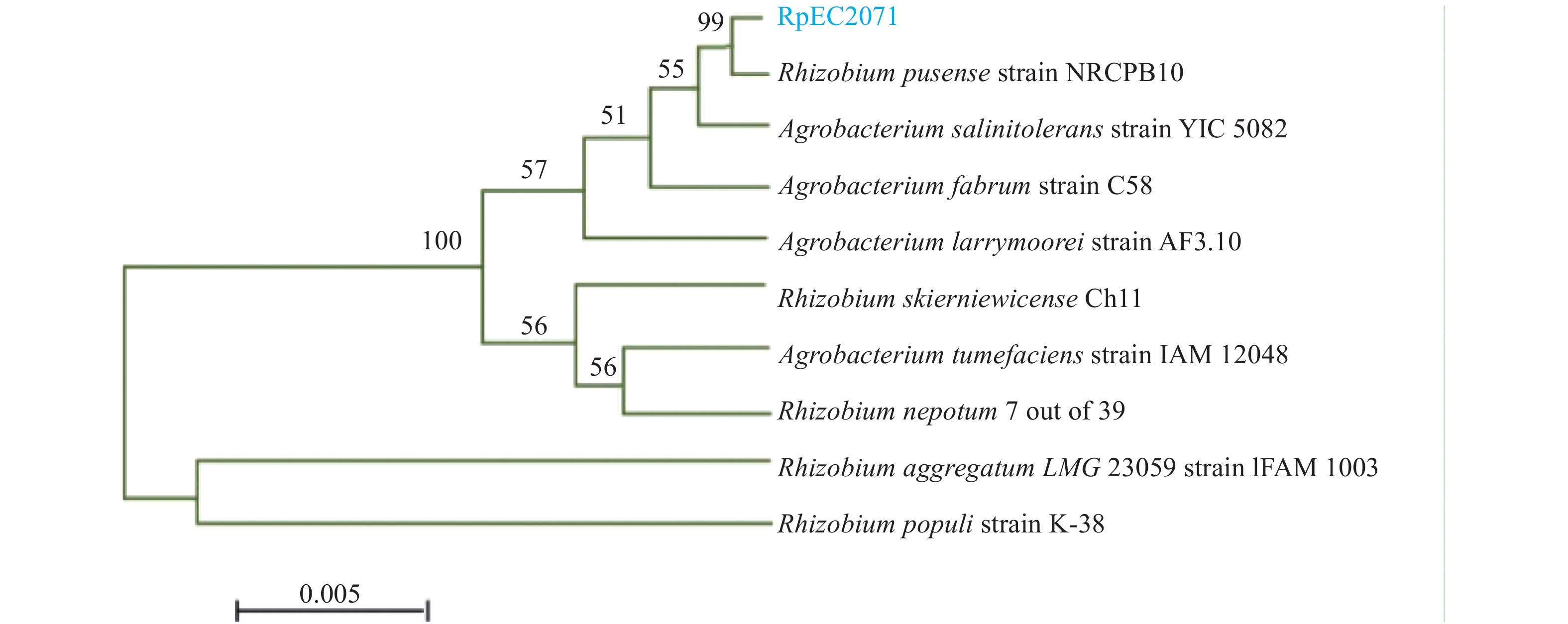
Biofilm-forming capacity of soil nitrogen-fixing bacteria and related gene expression under florfenicol combined with copper environmental stress
-
摘要:目的
研究氟苯尼考和铜(Cu)残留对土壤固氮菌的生态毒性效应,为评价兽药的环境风险提供依据。
方法对从花生根际土壤中分离出的一株优势固氮菌RpEC2071进行氟苯尼考和铜胁迫处理,试验设置空白组(0 μg/mL)、氟苯尼考添加组(40 μg/mL)、Cu添加组(200 μg/mL)和混合组(氟苯尼考40 μg/mL+Cu 200 μg/mL),并于给药后采样。采用苯酚硫酸法和96孔法研究氟苯尼考和Cu单独或联合处理对固氮菌胞外多糖产生及生物膜形成能力的影响。采用RT-qPCR测定分析固氮菌生物膜及固氮相关基因的mRNA表达水平。
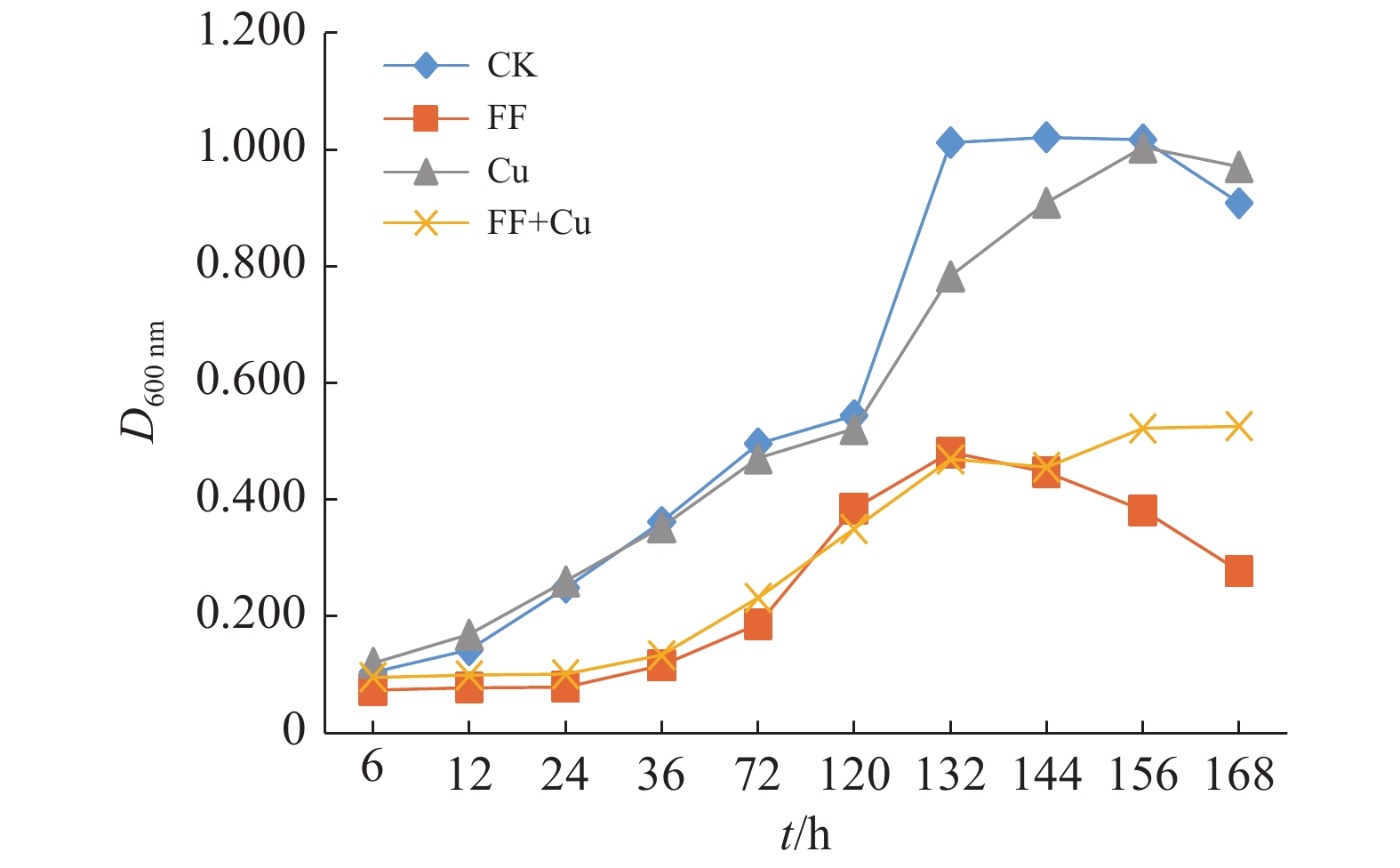
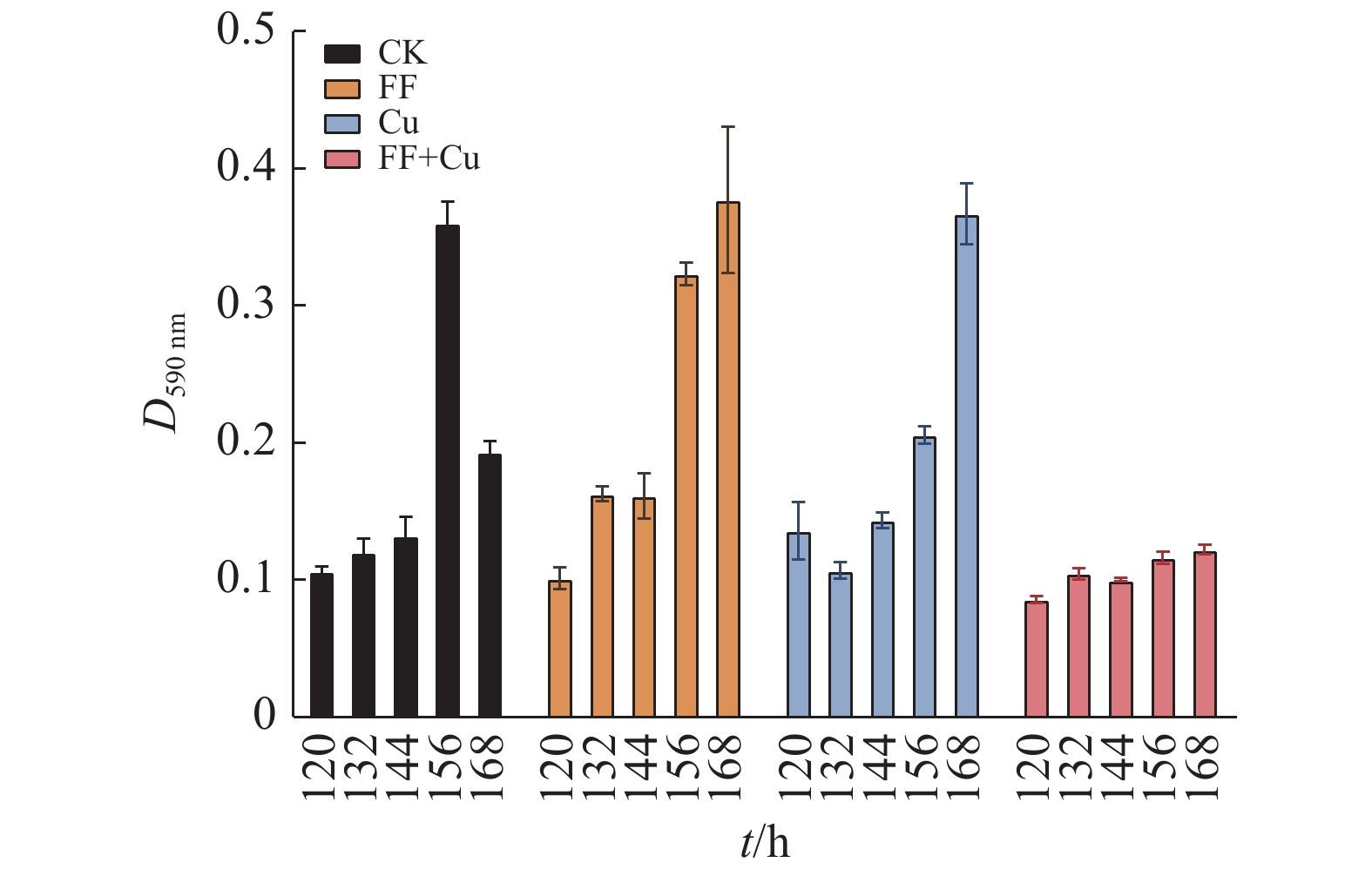
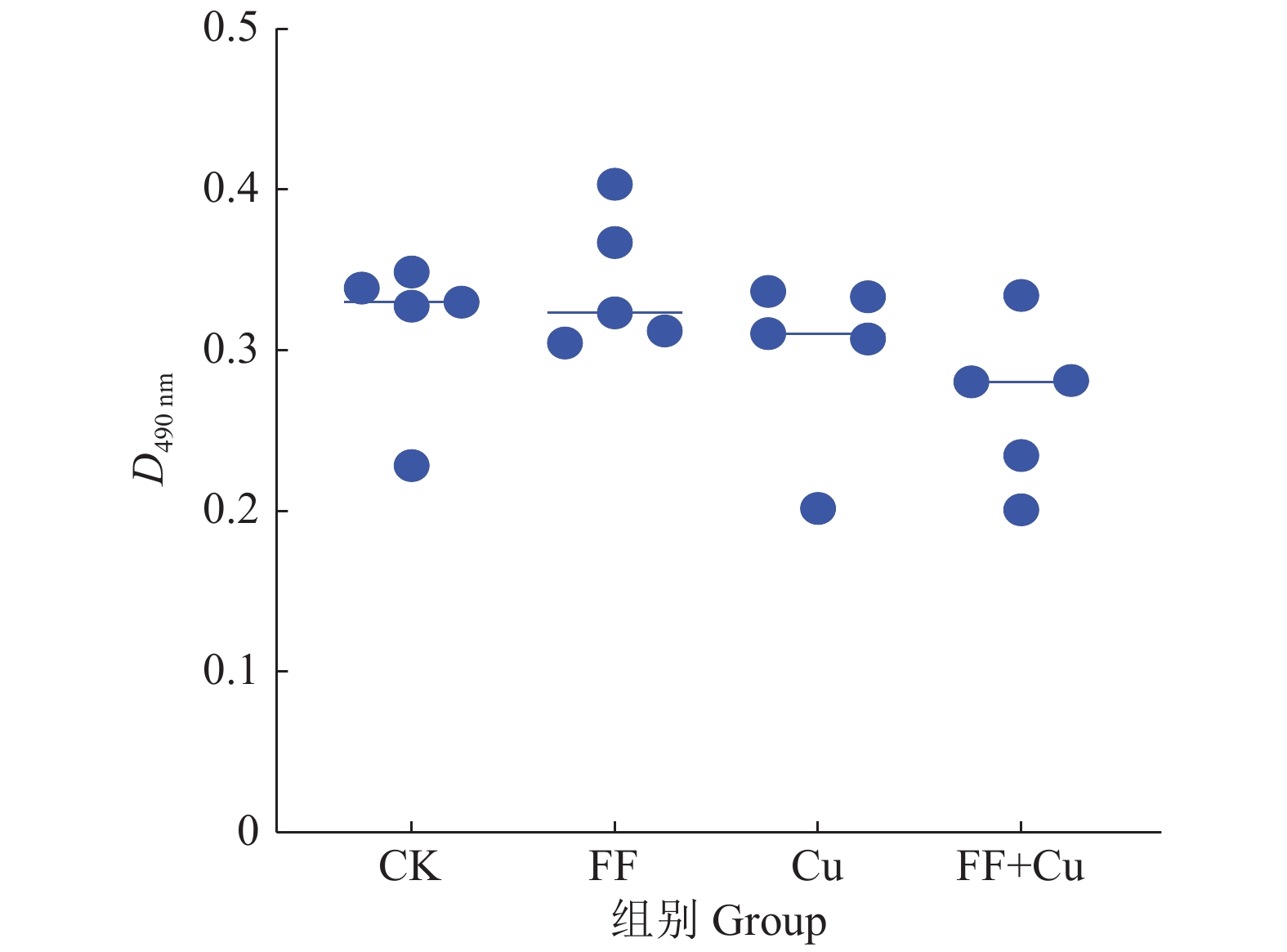
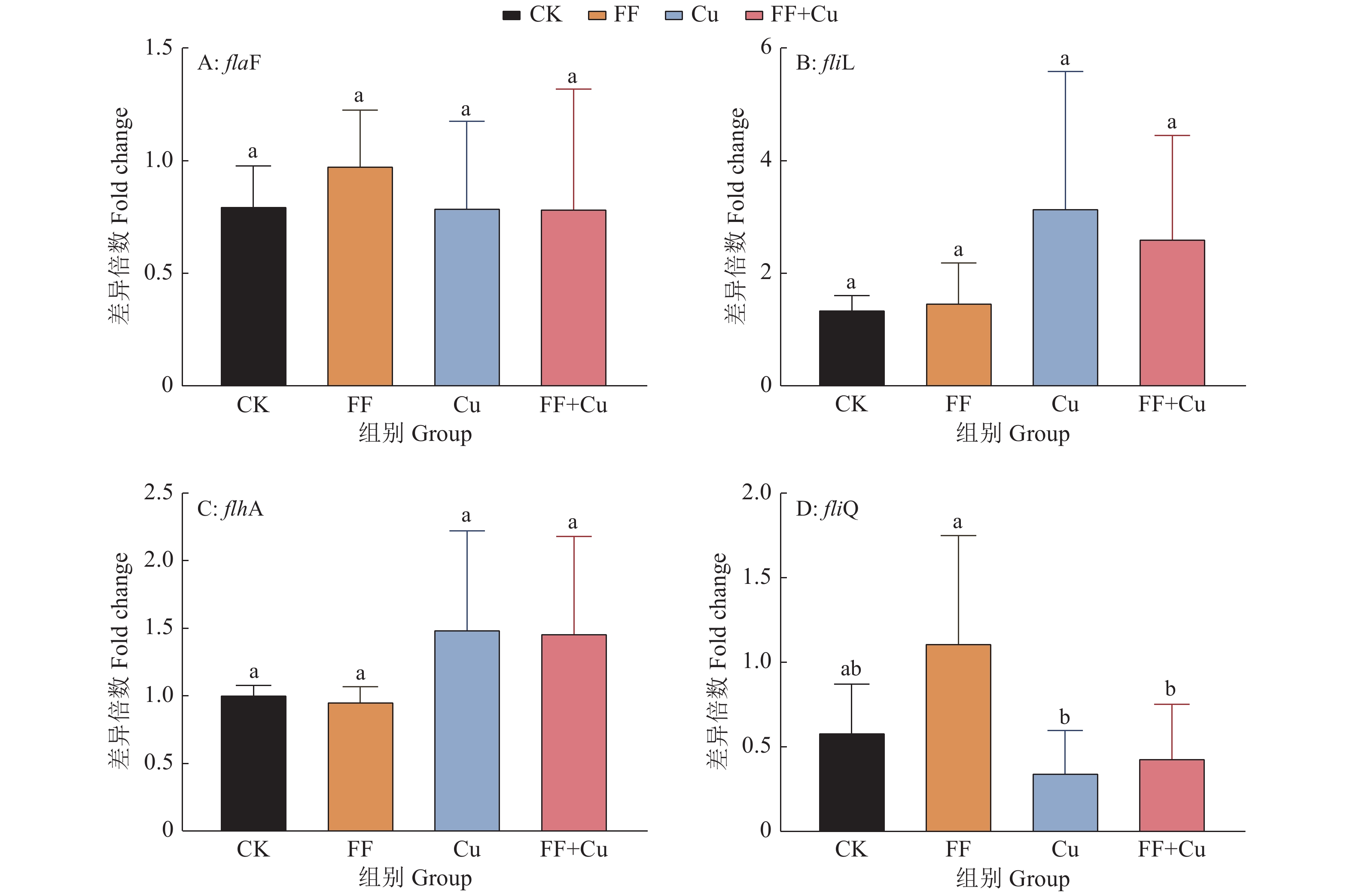
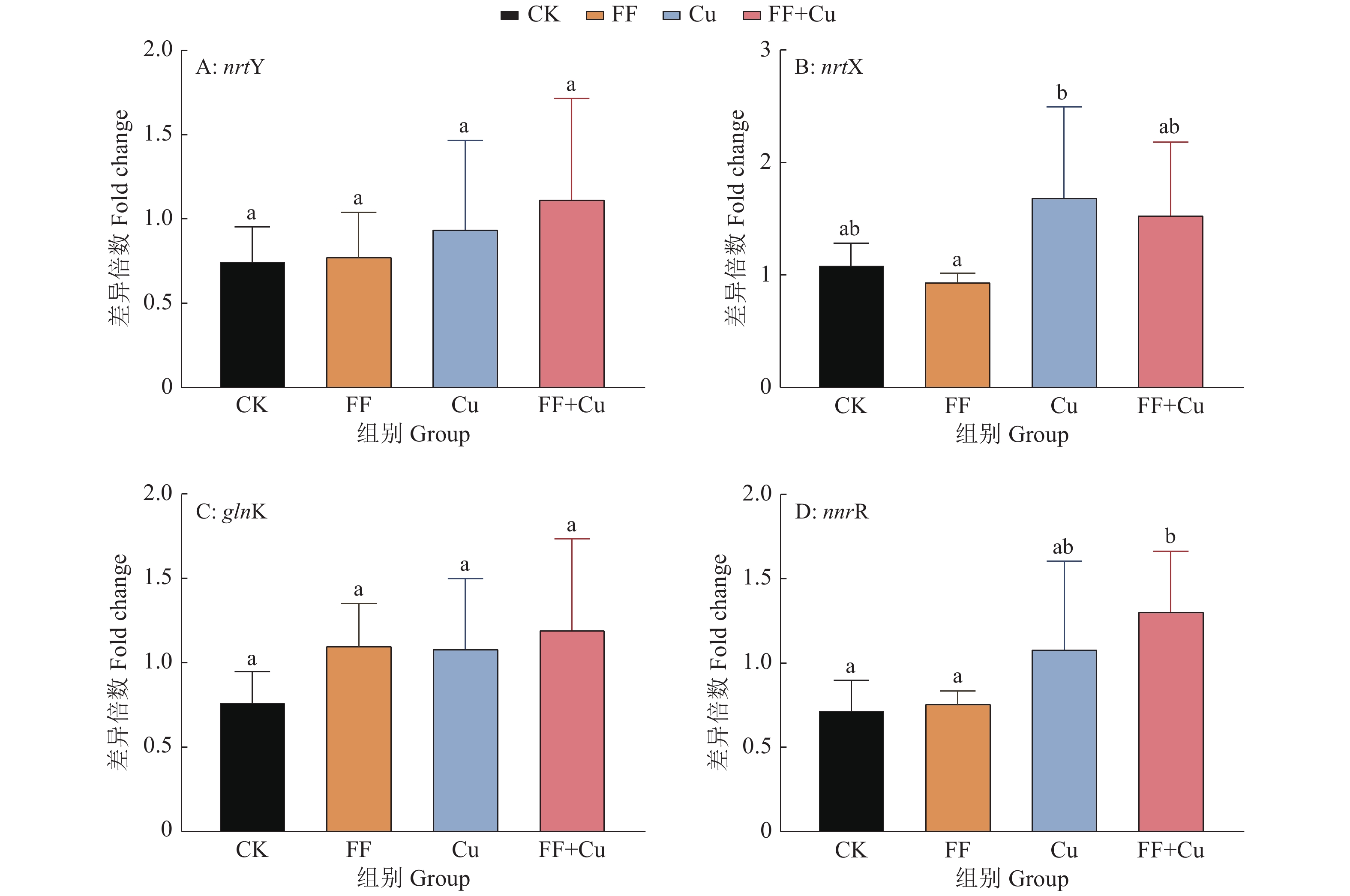
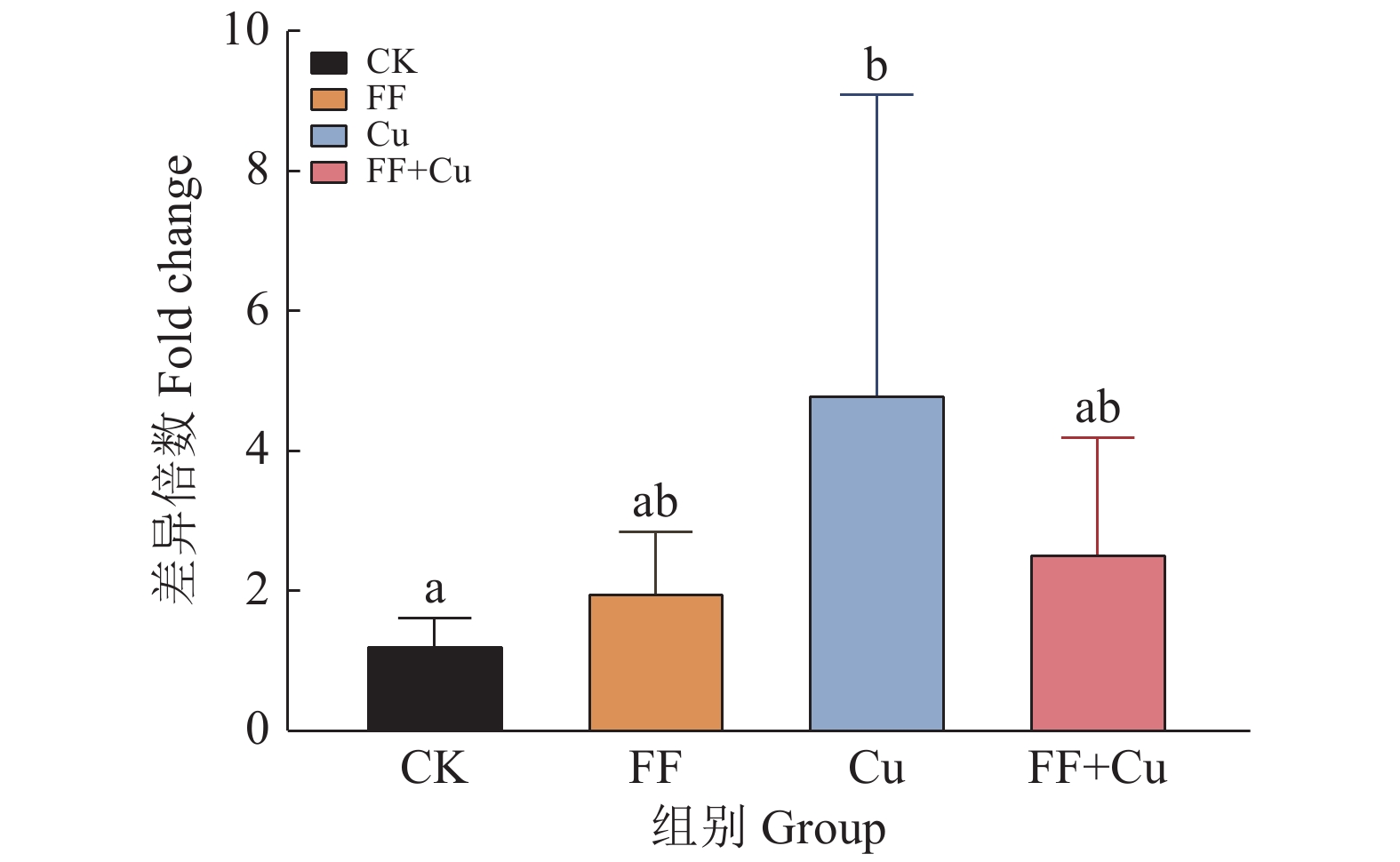
结果氟苯尼考和Cu单独胁迫会促进生物膜的形成,二者生物膜形成能力约为空白组的2倍;混合胁迫会抑制生物膜的形成,空白组生物膜形成能力是其3.1倍。胞外多糖分泌结果与生物膜形成能力测定结果基本一致。生物膜相关基因与氮代谢调控基因呈显著正相关,氟苯尼考和Cu的添加改变了fliQ、nifH和nnrR等基因的表达水平,且二者联合处理后会对其表达分别产生协同或拮抗作用。
结论氟苯尼考与Cu的单独或联合胁迫影响固氮菌RpEC2071中生物膜相关基因表达,进而对其生物膜形成能力产生影响。预示了以氟苯尼考和Cu为例的兽药及重金属残留胁迫会对土壤固氮菌产生潜在的生态毒性效应,从而破坏土壤中固氮生态系统。
Abstract:ObjectiveThe ecotoxic effects of florfenicol and copper (Cu) residues on soil nitrogen-fixing bacteria were studied to provide a basis for evaluating the environmental risks of veterinary drugs.
MethodA dominant nitrogen-fixing bacterium RpEC2071 was isolated from peanut root enclosure and treated under florfenicol and Cu stress. We set the blank group (0 μg/mL), florfenicol group (40 μg/mL), Cu group (200 μg/mL) and mixed group (flufenicol 40 μg/mL, Cu 200 μg/mL), and collected samples at multiple time points after dosing. The phenol-sulfuric acid method and 96-well microplate method were used to study the effects of florfenicol and Cu alone or in combination on the production of extracellular polysaccharides and biofilm formation of nitrogen-fixing bacterium. RT-qPCR was used to determine nitrogen-fixing bacterium biofilm formation and the mRNA expression levels of nitrogen fixation-related genes.
ResultFlorfenicol and Cu alone promoted the formation of biofilms, and the biofilm formation capacity of both was about twice that of the blank group. Under mixed stress, the biofilm formation was inhibited, and the biofilm formation capacity of the blank group was 3.1 times that of it. The results of extracellular polysaccharide secretion were basically consistent with the results of the determination of biofilm formation ability. Biofilm-related genes were significantly positively correlated with nitrogen metabolism regulatory genes, and the addition of florfenicol and Cu changed the expression levels of genes such as fliQ,ntrX andnnrR, and it would produce synergistic or antagonistic effects after the combination treatment of florfenicol and Cu.
ConclusionThe individual or combined stress of florfenicol and Cu affects the expression of biofilm-related genes in nitrogen-fixing bacteria RpEC2071, which in turn affects its ability to form biofilm. This research reveals the ecotoxic potential of veterinary drugs and heavy metal residue stresses on soil nitrogen-fixing bacteria, using florfenicol and Cu as examples. As a result, this can lead to the impairment of the nitrogen-fixing ecosystem in soil.
-
Keywords:
- Florfenicol /
- Copper /
- Nitrogen-fixing bacteria /
- Extracellular polysaccharide /
- Biofilm /
- Ecotoxicity
-
-
表 1 96孔板中处理组各组分添加量
Table 1 Addition of each component in a 96-well plate of treatment group
μL 组别 Group 改良肉汤 Improved broth 菌液 Bacterial solution 甲醇 Methanol 氟苯尼考1)Florfenicol CuSO42) 空白组 Blank(CK) 170 20 10 0 0 氟苯尼考组 Florfenicol(FF) 170 20 0 10 0 铜组 Cu(Cu) 160 20 10 0 10 混合组 Mixed(FF+Cu) 160 20 0 10 10 1) ρ (氟苯尼考) =0.8 mg/mL;2) ρ (CuSO4) = 4 mg/mL 1)ρ (Florfenicol) = 0.8 mg/mL; 2) ρ (CuSO4) = 4 mg/mL 表 2 胁迫模型组各组分添加量
Table 2 Addition of each component in stress model groups
mL 组别 Group 改良肉汤 Improved broth 菌液 Bacterial solution 甲醇 Methanol 氟苯尼考1)Florfenicol CuSO42) 空白组 Blank(CK) 40.1 0.1 0.1 0 0 氟苯尼考组 Florfenicol(FF) 40.1 0.1 0 0.1 0 铜组 Cu(Cu) 40.0 0.1 0.1 0 0.1 混合组 Mixed(FF+Cu) 40.0 0.1 0 0.1 0.1 1) ρ (氟苯尼考)=16 mg/mL;2) ρ (CuSO4)= 80 mg/mL 1) ρ (Florfenicol) = 16 mg/mL; 2) ρ (CuSO4) = 80 mg/mL 表 3 引物序列
Table 3 Primer sequences
基因名称 Gene name 引物序列(5′→3′) Primer sequence 基因名称 Gene name 引物序列(5′→3′) Primer sequence flaF F: GCGAGCGACAGGCGTTGA R: TGATTATCCGGCTGCTTGAGATC ntrX F: ACTTGTCGGTGCGTCACTTGC R: GATGGGCTTCTTCCAGTGCG fliL F: AAAACGAACAGGCAGAGGGC R: GGGAAACGGTGCGGACATAG glnK F: TGACCGTGACCGAAGTAAAGGG R: TGCCGTCGCCGATCTGCC flhA F: ACCACCAGTCATTTCCTTGCCC R: CCGCCGTCGGACCCTCAT nnrR F: GCTGGACGGATTGCTGACCC R: GCCACCCGACGCTCTACCTC fliQ F: ATCGTCGGTGTCGCCATCG R: CGTCATTTCCTGAACCTGCGTC nifH F: CGGATTATCGCAGTAGCAAACC R: TCTGTCGTCTCCATCGCTTCAC ntrY F: CGCTGACACCAAGTTCACGACG R: GCATCATGGAGTTCCAGATACCC Rp16S-1 F: AGATGCTCTACCTTGATGTCCCTG R: AGATGCGTTGCGCCACCT -
[1] 牛金利, 彭金菊, 明月月, 等. 氟苯尼考胁迫对土壤细菌耐药性的影响[J]. 中国兽医杂志, 2019, 55(4): 95-98. [2] CHOI J Y, KIM Y, KO E A, et al. Acinetobacter species isolates from a range of environments: Species survey and observations of antimicrobial resistance[J]. Diagnostic Microbiology and Infectious Disease, 2012, 74(2): 177-180. doi: 10.1016/j.diagmicrobio.2012.06.023
[3] 马驿, 彭金菊, 王芸, 等. 环丙沙星对土壤微生物量碳和土壤微生物群落碳代谢多样性的影响[J]. 生态学报, 2013, 33(5): 1506-1512. [4] 呼秀智, 薛占永, 王绥华, 等. 氟苯尼考对土壤微生物活动影响研究[J]. 山东畜牧兽医, 2011, 32(2): 3-5. [5] CAI L M, WANG Q S, LUO J, et al. Heavy metal contamination and health risk assessment for children near a large Cu-smelter in central China[J]. Science of the Total Environment, 2019, 650: 725-733. doi: 10.1016/j.scitotenv.2018.09.081
[6] 施昊坤, 吴次芳, 张茂鑫, 等. 土地整治对工业区周边土壤微生物多样性和群落结构影响分析[J]. 环境科学学报, 2020, 40(1): 212-223. [7] GAO M L, SONG W H, ZHOU Q, et al. Interactive effect of oxytetracycline and lead on soil enzymatic activity and microbial biomass[J]. Environmental Toxicology and Pharmacology, 2013, 36(2): 667-674. doi: 10.1016/j.etap.2013.07.003
[8] BAO Y Y, CHEN Q, MA W, et al. Influence of Fe addition on the accumulation of oxytetracycline in rice seedlings (Oryza sativa L. ) growing in hydroponic and soil culture[J]. Journal of Soils and Sediments, 2018, 18(5): 1958-1970. doi: 10.1007/s11368-018-1920-8
[9] WANG M, WU J, ZHOU T, et al. Effects of copper and florfenicol on nirS- and nirK-type denitrifier communities and related antibiotic resistance in vegetable soils[J]. Ecotoxicology and Environmental Safety, 2021, 213: 112011. doi: 10.1016/j.ecoenv.2021.112011
[10] FALKOWSKI P G. Volution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean[J]. Nature, 1997, 387(6630): 272-275. doi: 10.1038/387272a0
[11] 鲁文茹, 刘广锋, 郭志勋, 等. 金银花、连翘对溶珊瑚弧菌及其生物膜活性的影响[J]. 广东药学院学报, 2014, 30(3): 297-300. [12] 徐光域, 颜军, 郭晓强, 等. 硫酸−苯酚定糖法的改进与初步应用[J]. 食品科学, 2005, 26(8): 342-346. [13] 邹晓蕾, 刘礼崔, 罗立新. 细菌总RNA提取方法的比较[J]. 现代食品科技, 2013, 29(8): 1948-1954. [14] KARATAN E, WATNICK P. Signals, regulatory networks, and materials that build and break bacterial biofilms[J]. Microbiology and Molecular Biology Reviews, 2009, 73(2): 310-347. doi: 10.1128/MMBR.00041-08
[15] 冯世文, 曾芸, 李军, 等. 细菌对氟苯尼考的耐药机制研究[J]. 黑龙江畜牧兽医, 2014(5): 52-54. doi: 10.13881/j.cnki.hljxmsy.2014.0015 [16] 杜心恬, 宋馨, 刘欣欣, 等. 细菌胞外多糖生物合成转录调控因子研究进展[J]. 微生物学通报, 2021, 48(2): 573-581. [17] BOGINO P C, DE LAS MERCEDES OLIVA M, SORROCHE F G, et al. The role of bacterial biofilms and surface components in plant-bacterial associations[J]. International Journal of Molecular Sciences, 2013, 14(8): 15838-15859. doi: 10.3390/ijms140815838
[18] WANG D, XU A M, ELMERICH C, et al. Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions[J]. The ISME Journal, 2017, 11(7): 1602-1613. doi: 10.1038/ismej.2017.30
[19] LI S, PENG C, CHENG T, et al. Nitrogen-cycling microbial community functional potential and enzyme activities in cultured biofilms with response to inorganic nitrogen availability[J]. Journal of Environmental Sciences, 2019, 76: 89-99. doi: 10.1016/j.jes.2018.03.029
[20] YANG Z M, HAN Y L, MA Y, et al. Global investigation of an engineered nitrogen-fixing Escherichia coli strain reveals regulatory coupling between host and heterologous nitrogen-fixation genes[J]. Scientific Reports, 2018, 8(1): 10913-10928. doi: 10.1038/s41598-018-29241-9
[21] LI K H, WU G C, LIAO Y L, et al. RpoN1 and RpoN2 play different regulatory roles in virulence traits, flagellar biosynthesis, and basal metabolism in Xanthomonas campestris[J]. Molecular Plant Pathology, 2020, 21(7): 907-922. doi: 10.1111/mpp.12938
[22] CALATRAVA M, NOGALES J, AMEZTOY K, et al. The NtrY/NtrX system of Sinorhizobium meliloti GR4 regulates motility, EPS I production, and nitrogen metabolism but is dispensable for symbiotic nitrogen fixation[J]. Molecular Plant-Microbe Interactions, 2017, 30(7): 566-577. doi: 10.1094/MPMI-01-17-0021-R
[23] HAO J, FENG Y, WANG X, et al. Soil microbial nitrogen-cycling gene abundances in response to crop diversification: A meta-analysis[J]. Science of the Total Environment, 2022, 838(4): 156621. doi: 10.1016/j.scitotenv.2022.156621.
[24] BUENO E, ROBLES E, TORRES M, et al. Disparate response to microoxia and nitrogen oxides of the Bradyrhizobium japonicum napEDABC, nirK and norCBQD denitrification genes[J]. Nitric Oxide, 2017, 68: 137-149. doi: 10.1016/j.niox.2017.02.002
[25] BUENO E, MANIA D, MESA S, et al. Regulation of the emissions of the greenhouse gas nitrous oxide by the soybean endosymbiont Bradyrhizobium diazoefficiens[J]. International Journal of Molecular Sciences, 2022, 23(3): 1486. doi: 10.3390/ijms23031486.



 下载:
下载: