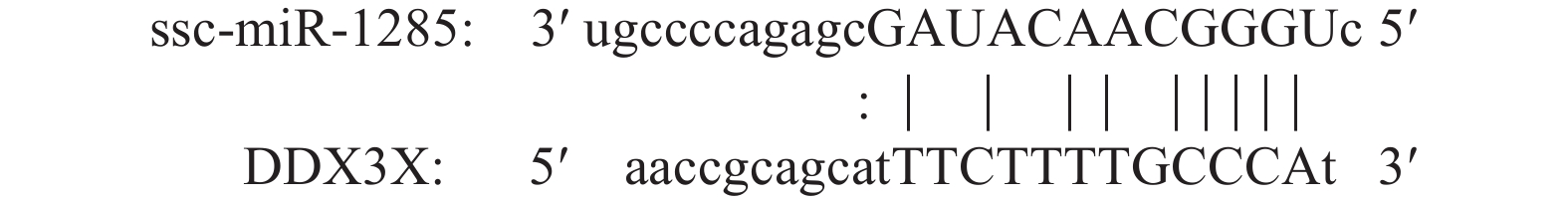Regulation effects of microRNA-1285 and its target DDX3X on Senecavirus A infected PK-15 cells
-
摘要:目的
探究MicroRNA-1285(miR-1285)及其靶标DDX3X在猪塞内卡病毒(Senecavirus A,SVA)感染PK-15细胞中的调控作用。
方法利用qRT-PCR、双荧光素酶活性及Western blot等方法研究miR-1285和DDX3X对I型干扰素(IFN-β)分泌及RIG-I信号通路的作用,分析miR-1285及DDX3X对SVA 3C蛋白基因表达的影响。
结果SVA感染PK-15细胞后,miR-1285表达量显著升高,并且miR-1285与DDX3X存在负靶向关系,二者可促进IFN-β转录及蛋白水平的表达。miR-1285通过靶向DDX3X对RIG-I信号通中的MAVS、TRAF3信号分子起调控作用。对于SVA 3C蛋白基因,DDX3X可以显著抑制其转录,并且可以逆转miR-1285所诱导的上调趋势。
结论SVA感染PK-15细胞后,宿主miR-1285及其靶标DDX3X对IFN-β及病毒3C蛋白的表达具有调控作用,研究结果将为明确miRNAs调控SVA感染的分子机制奠定基础,并为SVA的防控和诊断提供新的科学依据。
-
关键词:
- MicroRNA-1285 /
- DDX3X /
- 猪塞内卡病毒 /
- IFN-β /
- RIG-I信号通路
Abstract:ObjectiveTo explore the regulation roles of microRNA-1285 (miR-1285) and its target DDX3X in Senecavirus A (SVA) infected PK-15 cells.
MethodBy qRT-PCR, double luciferase activity and Western blot, the effects of miR-1285 and its target DDX3X on IFN-β secretion and the RIG-I signaling pathway were studied, and their effects on the expression of SVA 3C protein gene were analyzed.
ResultIn SVA infected PK-15 cells, the expression of miR-1285 increased significantly, and there was a negative targeting relationship between miR-1285 and DDX3X. Both miR-1285 and DDX3X promoted the transcription and protein expression of IFN-β. MiR-1285 regulated MAVS and TRAF3 signaling molecules in the RIG-I signaling pathway by targeting DDX3X. For SVA 3C protein, DDX3X significantly inhibited the transcription of 3C and reversed the up-regulation trend induced by miR-1285.
ConclusionAfter infecting PK-15 cells with SVA, host miR-1285 and its target DDX3X can regulate the expression of IFN-β and the viral 3C protein, which will lay a foundation for clarifying the molecular mechanism of miRNAs regulating SVA infection, and provide a new scientific basis for the prevention, control and diagnosis of SVA.
-
Keywords:
- MiRNA-1285 /
- DDX3X /
- Senecavirus A /
- IFN-β /
- RIG-I signal pathway
-
植物内生真菌(Endophytic fungus)是指那些生活史的部分阶段或全部生活于健康植物组织或器官内部,且在长期的协同进化过程中与宿主植物形成互惠共生关系的一类真菌[1-2]。内生真菌与宿主植物形成的这种互惠共生关系为研究者从植物内生真菌次生代谢产物中寻找新型的天然活性先导物提供了新的方向[3]。近年来,内生真菌次生代谢产物的研究越来越受到人们的重视,尤其是从短叶红豆杉Taxus brevifolia韧皮部分离到产紫杉醇的内生真菌Taxomyces andreanae后,掀起了人们对内生真菌次生代谢产物研究的热潮[4-5]。内生真菌作为一种新型的天然活性物质资源宝库,能够产生多种结构新颖的次生代谢产物,其具有抗菌、杀虫、抗氧化、抗肿瘤以及促进植物生长和提高植物抗病性等多种生物活性,从植物内生真菌中寻找具有开发潜能的生物活性物质引起了人们极大的兴趣[6-9]。
目前对于桉树内生真菌及其次生代谢产物的研究多集中在内生真菌的生理学和生态学作用方面。谢安强等[10-12]研究发现桉树内生真菌能够显著提高桉树的抗逆能力,如促进植株对磷的吸收能力,提高桉树的抗寒能力,促进植株的光合作用效能等。Kharwar等[13]从柠檬桉Eucalyptus citriodora中分离得到的曲霉属Aspergillussp.和毛壳菌属Chaetomiumsp.内生真菌表现出较强的抗真菌和抗细菌活性。格希格图等[14]从不同桉树根部分离到的内生真菌对桉树青枯病菌Ralstonia solanacearum有不同程度的拮抗作用,其中以大叶桉Eucalyptus robusta内生真菌的抗菌活性最为明显。Mohali等[15]从桉树和金合欢Acacia farnesiana中分离到2株新的内生真菌Fusicoccum sp.,但并未对其次生代谢产物的生物活性进行研究。华南农业大学林学与风景园林学院森林保护教研室的前期研究表明,窿缘桉Eucalyptus exserta果实内生真菌Eef-10表现出较好的抗桉树青枯病菌活性[16],本文在前期研究的基础上,以内生真菌Eef-10为研究对象,通过形态学和分子生物学相结合的方法确定其分类地位,同时分离和鉴定内生真菌Eef-10中的活性成分,并测定其抗细菌和抗肿瘤细胞活性,以期为内生真菌资源的开发和利用提供理论依据。
1. 材料与方法
1.1 供试菌株和肿瘤细胞
内生真菌Eef-10分离自健康的窿缘桉果实,目前保藏于华南农业大学林学与风景园林学院植物和微生物健康实验室。
供试细菌包括大肠埃希菌Escherichia coli(G-)、根癌土壤杆菌Agrobacterium tumefaciens(G-)、黄瓜角斑病菌Pseudomonas lachrymans(G-)、桉树青枯病菌Ralstonia solanacearum(G-)和番茄疮痂病菌Xanthomonas vesicatoria(G-),以上菌株均保藏于华南农业大学林学与风景园林学院植物和微生物健康实验室。
供试细胞株系为肝癌细胞(Hep-G2)和宫颈癌细胞(HeLa),供试肿瘤细胞由中国医学科学院北京协和医学院医药生物技术研究所提供。
1.2 仪器与试剂
核磁共振波谱仪(Bruker Avance-600,美国Bruker公司),半制备型液相色谱仪(泵:UC-3281,紫外检测器:UC-3292S,上海Welch公司),分析型液相色谱仪(LC-10F,天津博纳艾杰尔科技有限公司),三用紫外仪(ZQ-1,上海安亭科学仪器厂),超净工作台(SW-CJ-2G,苏州净化设备有限公司),生化培养箱(LRH-250,上海一恒科技有限公司)。
Sephadex LH-20葡聚糖凝胶(瑞士GE Heaithcare公司),GF254薄层层析硅胶和正相柱层析硅胶(青岛化工有限公司),硫酸链霉素(w=98%,上海麦克林生化科技有限公司),噻唑蓝(MTT)生物显色剂(w=98%,美国Amresco公司),喜树碱(w≥99%,上海麦克林生化科技有限公司),真菌基因组DNA抽提试剂盒 [生工生物工程(上海)股份有限公司],甲醇、二甲基亚砜、丙酮、乙酸乙酯、三氯甲烷、二氯甲烷和石油醚(分析纯,天津富宇精细化工厂),甲醇(色谱纯,上海星可高纯溶剂有限公司),氘代氯仿、氘代丙酮(巴斯夫化学有限公司)。
1.3 内生真菌Eef-10的鉴定
采用形态学和分子生物学相结合的方法对内生真菌Eef-10进行鉴定。
1.3.1 形态学鉴定
用灭菌牙签挑取PDA培养基上菌落边缘生长旺盛的菌丝,接种于新的PDA培养基中,置于生化培养箱中培养7~10 d,然后用生物显微镜观察菌丝、有性孢子和产孢结构等特征,参照Sekhar等[17]对内生真菌Eef-10进行形态学鉴定。
1.3.2 分子生物学鉴定
参照单体江等[18]的方法对内生真菌Eef-10进行分子生物学鉴定。将纯化后的内生真菌Eef-10(在PDA培养基上生长5 d)接种到PDB培养基中,28 ℃、150 r/min条件下振荡培养5 d,分液漏斗过滤后收集菌丝,用液氮进行充分研磨至粉末状,采用真菌基因组DNA抽提试剂盒提取其总DNA,使用真菌的通用引物ITS4(5′-TCCTCCGCTTATTGATATGC-3′)和ITS5(5′-GGAAGTAAAAGTCGTAACAAGG-3′)扩增其ITS序列。PCR反应体系(50 μL):去离子水21 μL,2×Taq PCR MasterMix(含染料)25 μL,ITS4(10 μmol/L)1 μL,ITS5(10 μmol/L)1 μL,模板DNA(10 ng/ μL)2 μL;PCR扩增程序:94 ℃预变性3 min;94 ℃变性40 s,56 ℃退火40 s,72 ℃延伸1 min 20 s,共30个循环;72 ℃延伸10 min。PCR产物送至生工生物工程(上海)股份有限公司进行测序。将测序成功的ITS序列提交至GenBank数据库,获得登录号。通过在NCBI网站上进行BLAST,下载与其相似性较高的序列及其近似属的序列,使用MAFTT version 7处理后,用MEGA6软件采用最大似然法构建系统发育树。
1.4 内生真菌Eef-10次生代谢产物的制备
采用大米固体培养基对内生真菌Eef-10进行大规模发酵。首先从4 ℃冻存管中挑取少许Eef-10菌丝接种到PDA平板上活化培养5~7 d,再将其菌丝接种在500 mL装有200 mL PDB培养基的三角瓶中,置于28 ℃、150 r/min条件下振荡培养3~5 d。将获得的液体种子接入已灭菌的大米培养基中,共发酵6 kg,28 ℃条件下暗培养60 d,以确保菌丝将大米培养基营养消耗完毕。而后将发酵好的固体发酵物用甲醇冷浸提取3次,每次7 d,减压浓缩后水混悬,依次用等体积的石油醚和乙酸乙酯萃取,分别得到石油醚层和乙酸乙酯层粗提物。
1.5 次生代谢产物的分离、纯化和鉴定
乙酸乙酯层提取物(51.40 g)经减压硅胶柱层析,先用石油醚洗脱,再用二氯甲烷和甲醇梯度洗脱,通过薄层层析合并成A~D 4个部分。其中B部分(25.19 g)再经减压硅胶柱层析,采用二氯甲烷和甲醇梯度洗脱,通过薄层层析合并成11个馏分,分别编号为B1~B11。B4馏分(2.9 g)经Sephadex LH-20柱层析(三氯甲烷︰甲醇体积比为1︰1),通过薄层层析检测后合并为7个组分,分别编号为B4-1~B4-7。B4-7(30 mg)经semi-HPLC(甲醇︰水体积比为 65︰35,流速5 mL/min,λ = 210 nm,进样体积为0.1 mL)制备,得到化合物Ⅰ(4.5 mg)和化合物Ⅱ(5.5 mg)。B6馏分(9.5 g),经Sephadex LH-20(三氯甲烷︰甲醇体积比为1︰1)柱层析,通过薄层层析检测后合并成6个组分,编号为B6-1~B6-6。其中B6-3(95 mg)经semi-HPLC(甲醇︰水体积比为50︰50,流速5 mL/min,λ = 210 nm,进样体积为0.1 mL)制备,得到化合物Ⅲ(30 mg)。
单体化合物的结构主要通过1H NMR、13C NMR等波谱学数据进行鉴定,并与文献比对后确定。
1.6 抗细菌活性的测定
抗细菌活性的测定参照刘志强等[19]的方法,精密称取单体化合物Ⅰ~Ⅲ各2.0 mg分别溶于0.3 mL丙酮中,再加入0.7 mL蒸馏水,配成质量浓度为2 000 μg/mL的母液,然后用φ为30%的丙酮溶液依次稀释成质量浓度为2 000.000、1 000.000、500.000、250.000、125.000、62.500、31.250、15.625 μg/mL的样品溶液。阳性对照为硫酸链霉素,采用相同的方法配成质量浓度为500.000 00、250.000 00、125.000 00、62.500 00、31.250 00、15.625 00、7.812 50、3.906 25 μg/mL的溶液,备用。在无菌的96微孔板中加入106 CFU/mL的供试菌液90 μL,然后加入不同浓度的测试样品溶液10 μL。同时设空白对照(H2O)和溶剂对照(φ为30%的丙酮溶液),每处理6个重复。用封口膜将96微孔板四周封口,于28 ℃、黑暗条件下振荡(15 r/min)培养24 h,每孔加入10 μL MTT溶液(5 mg/mL),继续培养4 h后,以3 000 r/min 离心20 min,去上清,每孔加入二甲基亚砜(DMSO)150 μL,振荡(15 r/min)30 min,为了测定半抑制浓度(IC50),多孔板离心后,每孔取100 μL,于510 nm 下测定吸光度(D510 nm)。按下面公式计算待测样品对供试细菌的抑制率:
$${\text{抑制率}} = \frac{{{\text{溶剂对照孔}}{D_{510\;{\rm{nm}}}} - {\text{药液孔}}{D_{510\;{\rm{nm}}}}}}{{{\text{溶剂对照孔}}{D_{510\;{\rm{nm}}}}}} \times 100\% {\text{。}}$$ 所得数据采用 Microsoft excel 软件进行分析,供试样品浓度取对数(X),抑制率换算成概率值(Y),求得抑制活性回归方程(Y = aX+b)和半抑制浓度(IC50)。
1.7 抗肿瘤细胞活性测定
抗肿瘤细胞活性的测定参照Ouyang等[20]的方法,阳性对照为喜树碱。将对数生长期细胞接种于96孔细胞培养板(Nest)中,待6~8 h后细胞贴壁。用含有5%(w)FBS(Gibco)的DMEM(Hyclone)细胞培养基将化合物以2倍梯度稀释,共制备8个梯度(质量浓度分别为80.000、40.000、20.000、10.000、5.000、2.500、1.250和0.625 μg/mL)备用。小心吸弃各孔内原生长培养基后,将制备好的各浓度化合物分别加入对应孔中,每孔200 μL,每个化合物每个浓度设置4组重复孔。对照孔加入相应浓度的DMSO。在37 ℃,φ(CO2)为5%条件下,化合物与细胞继续共培养48 h。每孔加入CCK8试剂15 μL,在37 ℃、φ(CO2)为5%条件下继续培养4 h后,采用多功能微孔板检测仪在450 nm下检测各孔的吸光度(D450 nm),记录并保存数据。采用GraphPad Prism 5拟合各化合物在各细胞株的生长抑制曲线并计算其IC50值。
2. 结果与分析
2.1 内生真菌Eef-10的鉴定
内生真菌Eef-10的菌落和显微形态如图1所示。在PDA培养基上,菌落呈规则圆形(图1a),气生菌丝发达,绒毛状,初始时菌落白色,后期菌落颜色逐渐变灰,且随时间延长而逐渐加深,培养7 d后可布满整个培养皿(d = 7.5 cm)。在显微镜下观察发现,菌丝有隔(图1b),内含多个油滴,菌丝后期颜色较深且粗细不均,隔间距缩短;子囊果近球形(图1d),有孔口;附属丝较长,呈菌丝状,有隔,周生于子囊果且大部分集中生长于子囊果顶部;子囊孢子灰褐色(图1c),单孢,卵圆形,光滑,长×宽平均为7.0 μm×5.5 μm,有顶生萌发孔;与毛壳菌属Chaetomium的特征相一致[17, 21]。
进一步通过真菌通用引物ITS4和ITS5进行PCR扩增,获得大小为592 bp的目的序列,将该目的序列提交至GenBank,获得登录号MK120863。在GenBank数据库中进行同源性比对,采用最大似然法通过MEGA 6构建系统发育树(图2)。从图2可以看出,内生真菌Eef-10与Chaetomium sp.(登录号:KU504294.1)聚在同一支上,其最大相似度为99.82%,结合形态学特征最终将内生真菌Eef-10确定为子囊菌门Ascomycota核菌纲Pyrenomycetes粪壳目Sordariales毛壳菌科Chaetomiaceae毛壳菌属Chaetomium sp.真菌。
2.2 化合物的结构鉴定
通过1H NMR、13C NMR等波谱学数据以及与文献比对,确定了3种化合物的结构(图3)。
化合物Ⅰ,淡黄色固体,1H NMR谱在δH6.20 (1H, s)处有1个苯基质子信号,在δH2.09 (3H, s)和2.46 (3H, s)处有2个苯甲基质子信号,在δH3.91 (3H, s)处有1个甲氧基质子信号,δH 12.06 (1H, s)和 5.14 (1H, s)表明其结构中含有2个羟基信号,而且13C NMR谱中的2个芳香族碳信号δC163.37和158.20进一步支持了苯环在C-2和C-4处的氧化;通过1H NMR谱的相关信息及13C NMR谱δC163.37,158.20,140.39,110.75,108.70,105.46可以推导其结构中含有1个五取代苯环;13C NMR谱在δ 172.83处表明其结构中含有1个羰基。化合物Ⅰ的1H NMR (600 MHz,CDCl3)δH:12.06 (1H,s,2-OH),6.20 (1H,s,H-5),5.14 (1H,s,5-OH),3.91 (3H,s,H-1'),2.45 (3H,s,H-6),2.10 (3H,s,H-3);13C NMR (151 MHz,CDCl3) δC:172.83 (C-7),163.37 (C-2),158.20 (C-4),140.39 (C-6),110.75 (C-5),108.70 (C-3),105.46 (C-1),52.08 (C-1'),24.36 (C-6),7.89 (C-3)。上述数据与文献[22]数据一致,故将化合物Ⅰ鉴定为2, 4−二羟基−3, 6−二甲基苯甲酸甲酯(Methyl 2, 4-dihydroxy-3, 6-dimethylbenzoate,即Atraric acid)(图3a)。
化合物Ⅱ,白色固体,化合物Ⅱ的1H NMR谱和13C NMR谱与化合物Ⅰ数据非常接近,基本骨架一致,1H NMR谱中δH1.40 (t,J = 7.1 Hz,3H),4.38 (q,J = 7.1 Hz,2H)信号,表明其结构中含有1个R—CH2—CH3信号。化合物Ⅱ的1H NMR (600 MHz,CDCl3)δH:12.14 (1H,s,2-OH),6.19 (1H,s,H-5),5.18 (1H,s,5-OH),4.38 (2H,q,J= 7.1 Hz,H-1′),2.46 (3H,s,H-6),2.09 (3H,s,H-3),1.40 (3H,t,J= 7.1 Hz,H-2′);13C NMR (151 MHz,CDCl3) δC:172.17 (C-7),163.18(C-2),158.01 (C-4),140.18 (C-6),110.53 (C-5),108.55 (C-3),105.28(C-1),61.22(C-1′),24.23(C-6),14.26(C-2′),7.67(C-3)。上述数据与文献[23]数据一致,最终化合物Ⅱ鉴定为2, 4−二羟基−3, 6−二甲基苯甲酸乙酯(Ethyl 2, 4-dihydroxy-3, 6-dimethylbenzoate)(图3b)。
化合物Ⅲ,无色油状液体,化合物Ⅲ的1H NMR (600 MHz,Acetone-d6) δH:5.71(1H, s,H-3),4.32 (2H,t,J = 6.2 Hz,H-6),2.42 (2H,t,J = 6.2 Hz,H-5),2.01(3H,s,H-7);13C NMR (151 MHz,Acetone-d6)δC:164.55 (C-2),159.51 (C-4),116.95 (C-3),66.64 (C-6),29.75 (C-5),22.87(C-7)。上述数据与参考文献[24]数据基本一致,所以化合物Ⅲ鉴定为4−甲苯−5, 6−二氢−2H−吡喃−2−酮(4-methyl-5, 6-dihydro-2H-pyran-2-one)(图3c)。
2.3 抗细菌活性
内生真菌Eef-10次生代谢产物对5种供试细菌的抑制活性如表1所示。其中化合物Ⅲ在供试浓度下对5种供试细菌的IC50均大于200 μg/mL;化合物Ⅱ对所有供试细菌的抑制活性均强于化合物Ⅰ,其中对桉树青枯病菌的抑制活性最强,其IC50为35.87 μg/mL,与阳性对照硫酸链霉素非常接近,其次是对黄瓜角斑病菌和番茄疮痂病菌的抑制活性,其IC50分别为38.91和 40.80 μg/mL,对大肠埃希菌和根癌土壤杆菌的抑制活性较弱;化合物Ⅰ对黄瓜角斑病菌的抑制活性最强,其IC50为67.25 μg/mL,其次是对番茄疮痂病菌的抑制活性,其IC50为93.59 μg/mL,而对其他供试细菌的IC50均大于100 μg/mL。
表 1 内生真菌Eef-10次生代谢产物的抗细菌活性Table 1. Antibacterial activities of the secondary metabolites isolated from endophytic fungus Eef-10供试样品
Tested sampleIC50/(μg·mL−1) 大肠埃希菌
Escherichia
coli根癌土壤杆菌
Agrobacterium
tumefaciens黄瓜角斑病菌
Pseudomonas
lachrymans桉树青枯病菌
Ralstonia
solanacearum番茄疮痂病菌
Xanthomonas
vesicatoria化合物Ⅰ Compound Ⅰ 130.55 ± 3.57 105.39 ± 4.52 67.25 ± 1.23 113.11 ± 3.58 93.59 ± 0.43 化合物Ⅱ Compound Ⅱ 48.52 ± 0.33 55.50 ± 1.61 38.91 ± 0.54 35.87 ± 0.18 40.80 ± 0.70 化合物Ⅲ Compound Ⅲ > 200 > 200 > 200 > 200 > 200 硫酸链霉素 Streptomycin sulfate 18.51 ± 0.46 5.10 ± 0.03 30.54 ± 0.89 33.07 ± 2.46 13.81 ± 1.62 2.4 抗肿瘤细胞活性
抗肿瘤细胞活性的测定结果如表2所示,通过GraphPad Prism 5拟合各化合物在各细胞株的生长抑制曲线计算其IC50,除化合物Ⅱ之外,其他化合物对2种肿瘤细胞的IC50均大于50 μg/mL;化合物Ⅱ对HeLa细胞的IC50大于50 μg/mL,但对Hep-G2的IC50仅为1.50 μg/mL,明显强于阳性对照喜树碱的3.6 μg/mL,表明化合物Ⅱ对Hep-G2肿瘤细胞具有较好的生长抑制作用。
表 2 内生真菌Eef-10次生代谢产物的抗肿瘤细胞活性Table 2. Antitumor activities of the secondary metabolites isolated from endophytic fungus Eef-10供试细胞Tested cell IC50/(μg·mL−1) 化合物Ⅰ
Compound Ⅰ化合物Ⅱ
Compound Ⅱ化合物Ⅲ
Compound Ⅲ喜树碱
CamptothecinHep-G2 > 50 1.50 > 50 3.6 HeLa > 50 > 50 > 50 6.3 3. 讨论与结论
本研究以分离和鉴定抗细菌和抗肿瘤的活性化合物为导向,从窿缘桉内生真菌Chaetomium sp. Eef-10中共分离出3个化合物,包括2个苯酚类和1个戊烯酸内酯类化合物。前人的研究表明,化合物Ⅰ(Atraric acid)是一种天然的雄激素受体拮抗剂,主要应用于前列腺癌的治疗[25-26],该化合物曾在地衣植物Parmotrema cooperi[27]、Lecidella carpathica[28]和Pseudevernia furfuracea[22]以及地钱植物Frullania brasiliensis[29]、非洲臀果木Pygeum africanum[23]和紫葳科植物Newbouldia laevis[30]中被发现和报道,而有关化合物Ⅱ的研究却很少,一般是作为Atraric acid衍生物的形式被报道,但化合物Ⅱ对雄激素受体的拮抗作用甚至略高于Atraric acid,亦可作为抗前列腺癌优先级的候选药物[31]。本研究中活性测定的结果显示,化合物Ⅱ表现出较好的抗细菌活性,其中对桉树青枯病菌的抑制活性最强,IC50为35.87 μg/mL,与阳性对照结果相当,且化合物Ⅱ对Hep-G2肿瘤细胞也具有较好的抑制作用,研究结果为寻找抗桉树青枯病菌天然微生物源活性药物提供了新的思路。
致谢:感谢华南农业大学农学院谢辉和徐春玲老师在内生真菌形态学鉴定方面提供的帮助!
-
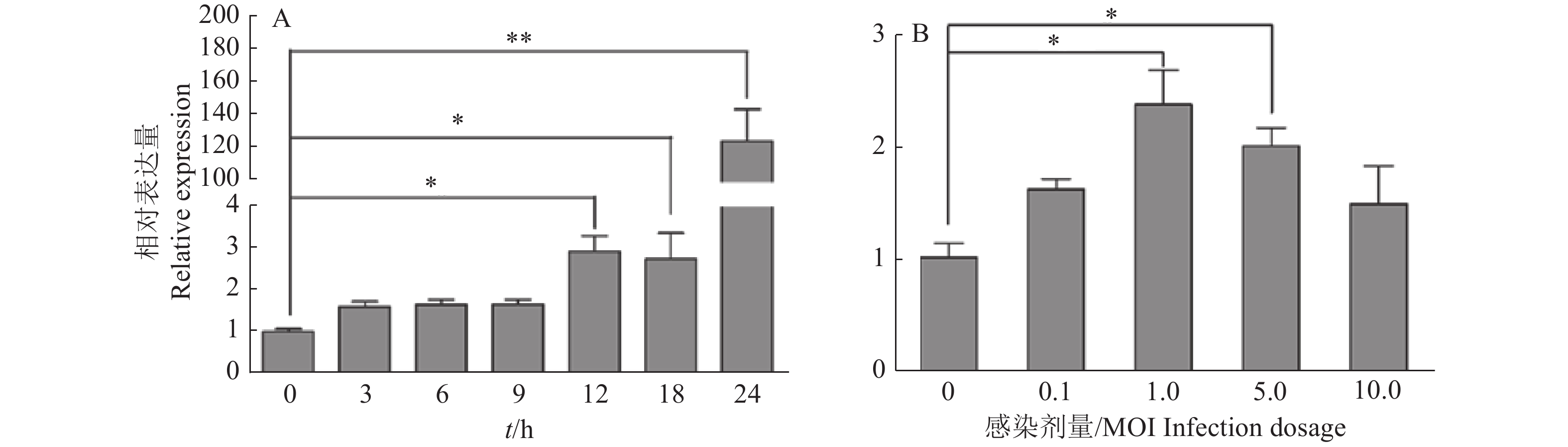
图 1 不同SVA感染时间(A)和感染剂量(B)条件下PK-15细胞中miR-1285的表达量
“*”“**”分别表示处理与对照在P < 0.05和P < 0.01水平差异显著(Duncan’s法)
Figure 1. Expression of miR-1285 in PK-15 cells infected by SVA at different infection time (A) and dosages (B)
“*” and “**” represented statistical difference in comparison with control group at P < 0.05 and P < 0.01 levels respectively (Duncan’s method)
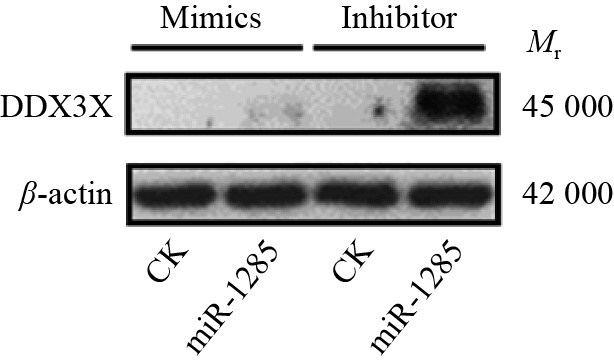
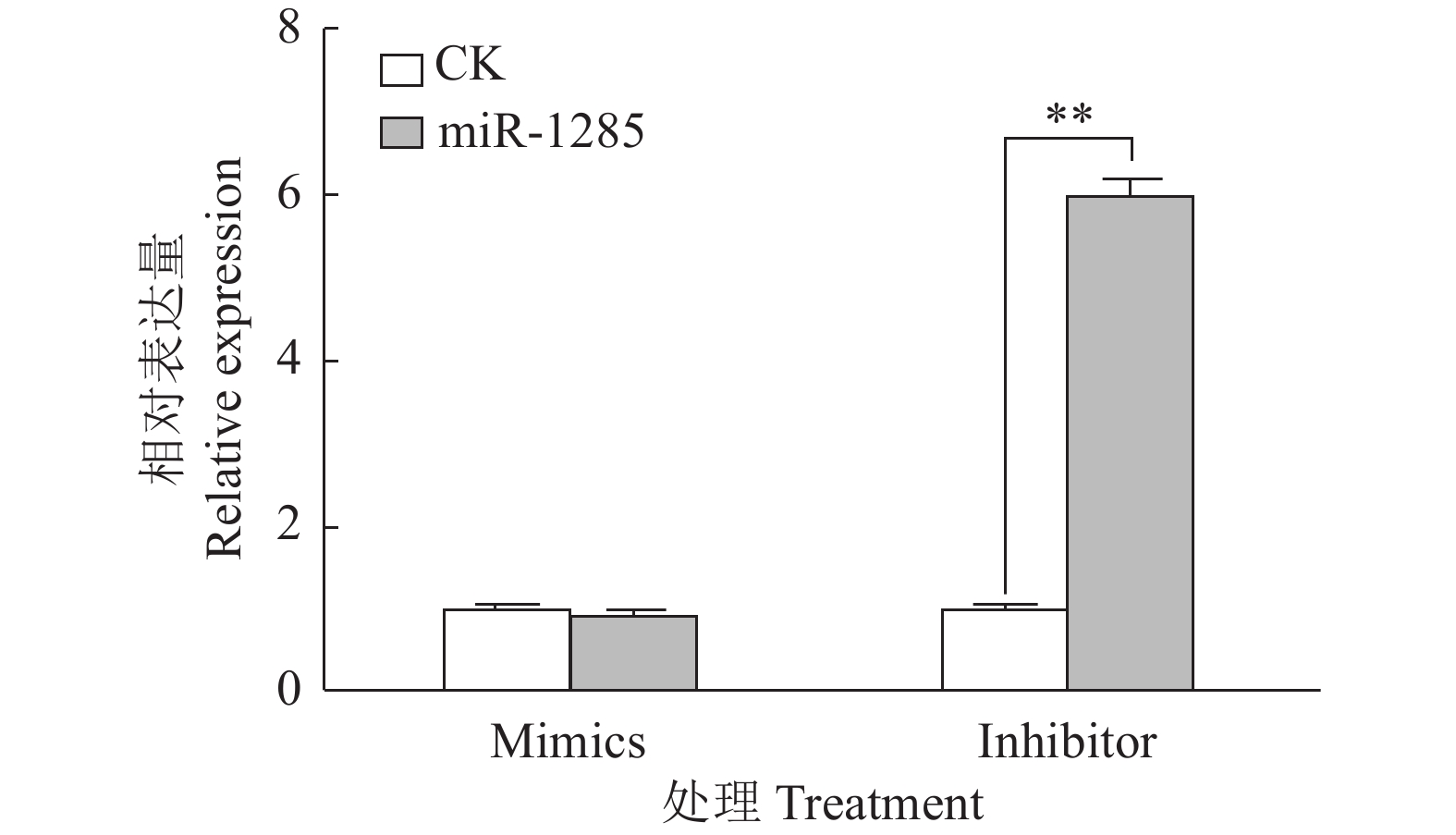
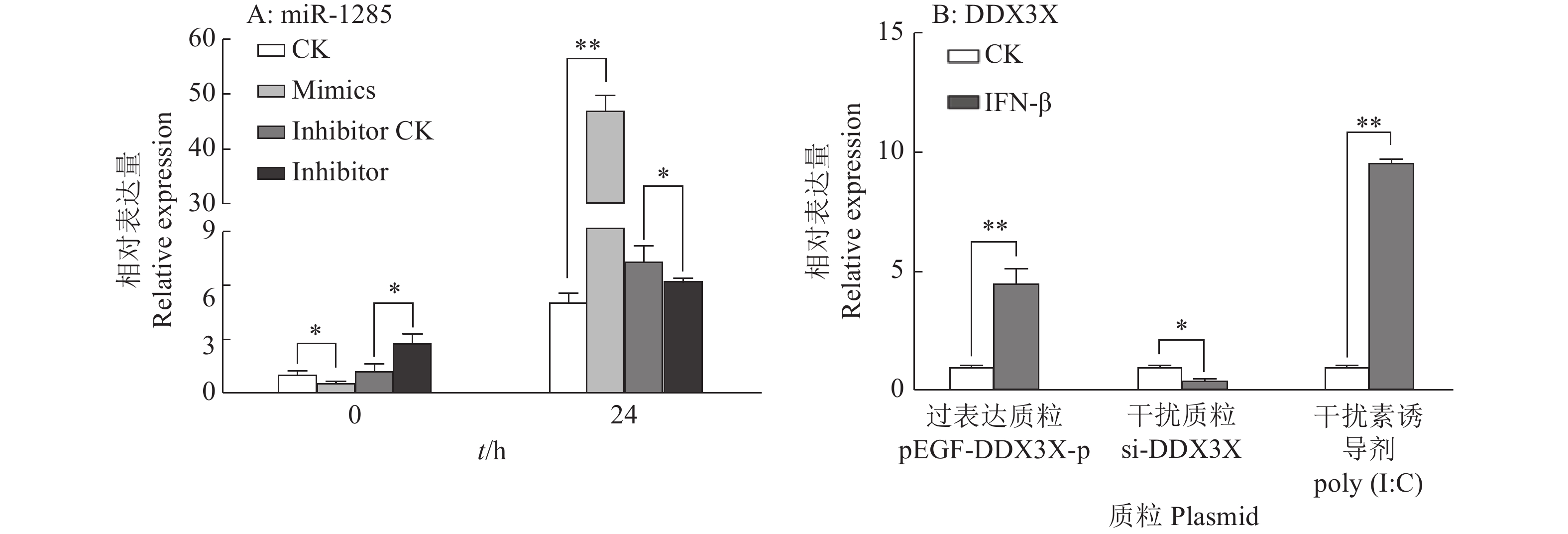
图 4 转染miR-1285 mimics、inhibitor至PK-15细胞后DDX3X的mRNA相对表达量
“**”表示处理与对照在P < 0.01水平差异显著 (Duncan’s 法)
Figure 4. The relative expression of DDX3X mRNA after transfection of miR-1285 mimics and inhibitor into PK-15 cells
“**” represented statistical difference in comparison with control group at P < 0.01 level (Duncan’s method)
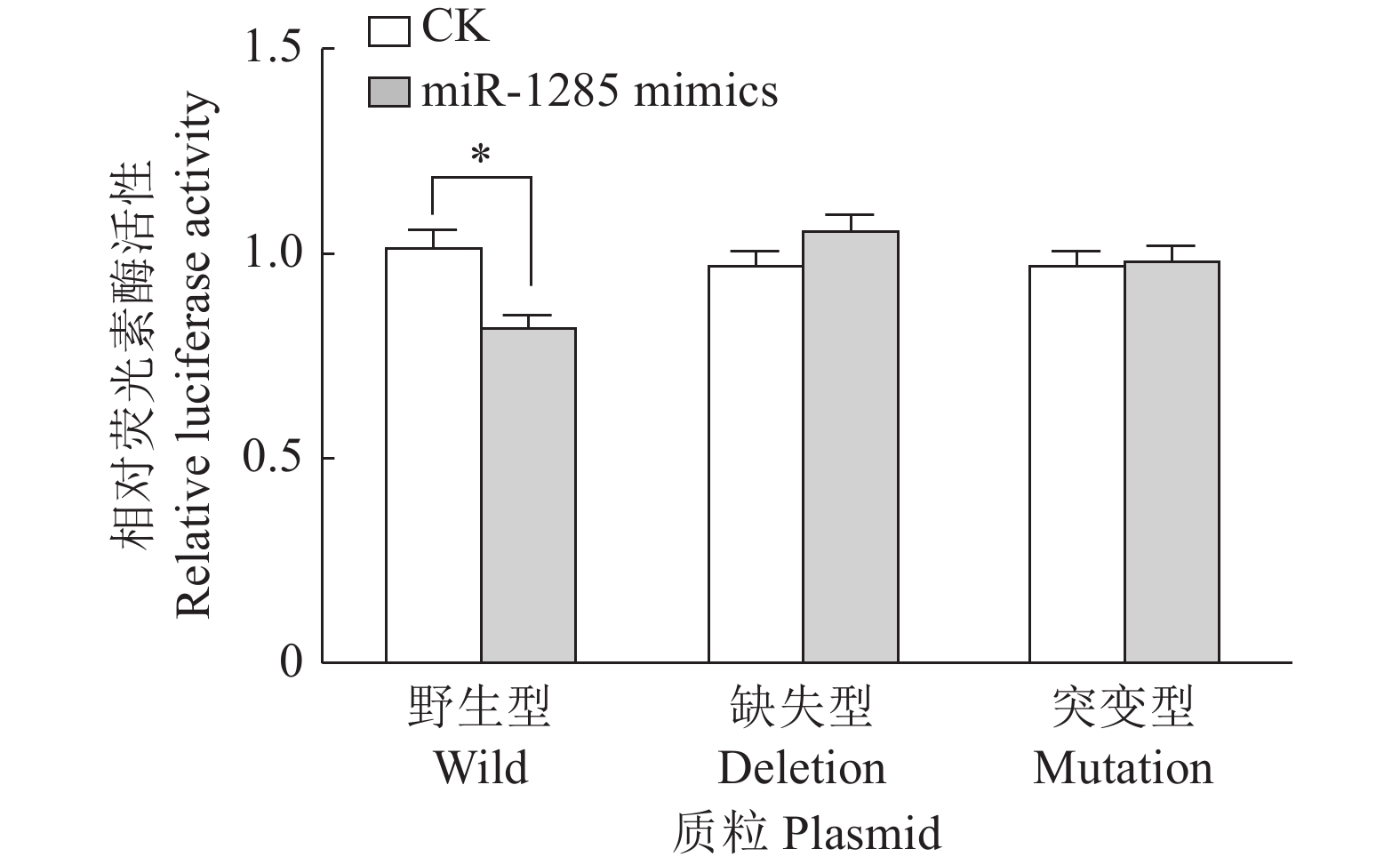
图 5 转染不同DDX3X重组载体质粒至PK-15细胞后miR-1285双荧光素酶活性
“*”表示处理与对照在P < 0.05水平差异显著 (Duncan’s 法)
Figure 5. The relative dual-luciferase activity of miR-1285 after transfection of different DDX3X recombinant vector plasmids into PK-15 cells
“*” represented statistical difference in comparison with control group at P < 0.05 level (Duncan’s method)
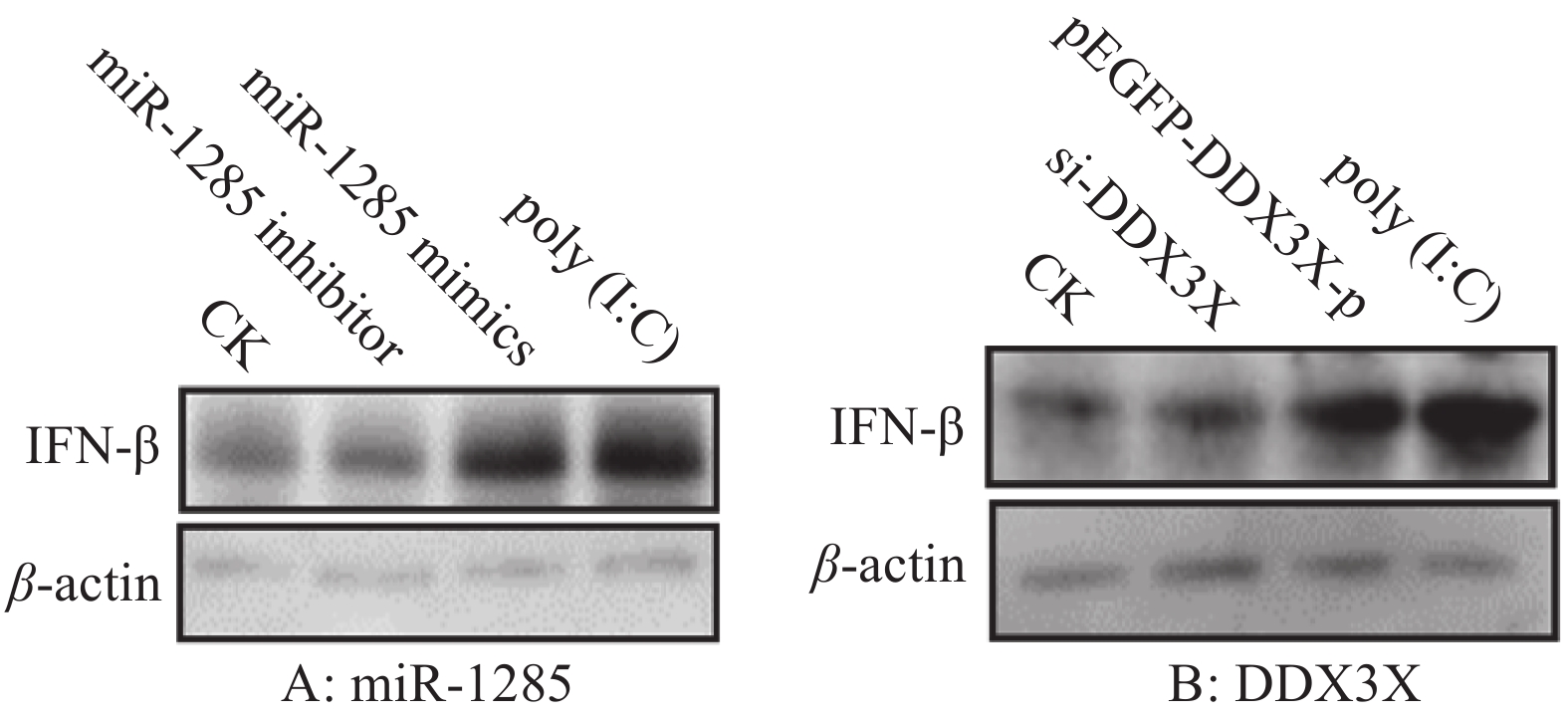
图 6 miR-1285及其靶标DDX3X对IFN-β mRNA表达的调控作用
“*”和“**”分别表示处理与对照在P < 0.05和P < 0.01水平差异显著(Duncan’s法)
Figure 6. Regulation effects of miR-1285 and its target DDX3X on the mRNA expression of IFN-β
“*” and “**” represented statistical difference in comparison with control group at P < 0.05 and P < 0.01 levels respectively (Duncan’s method)
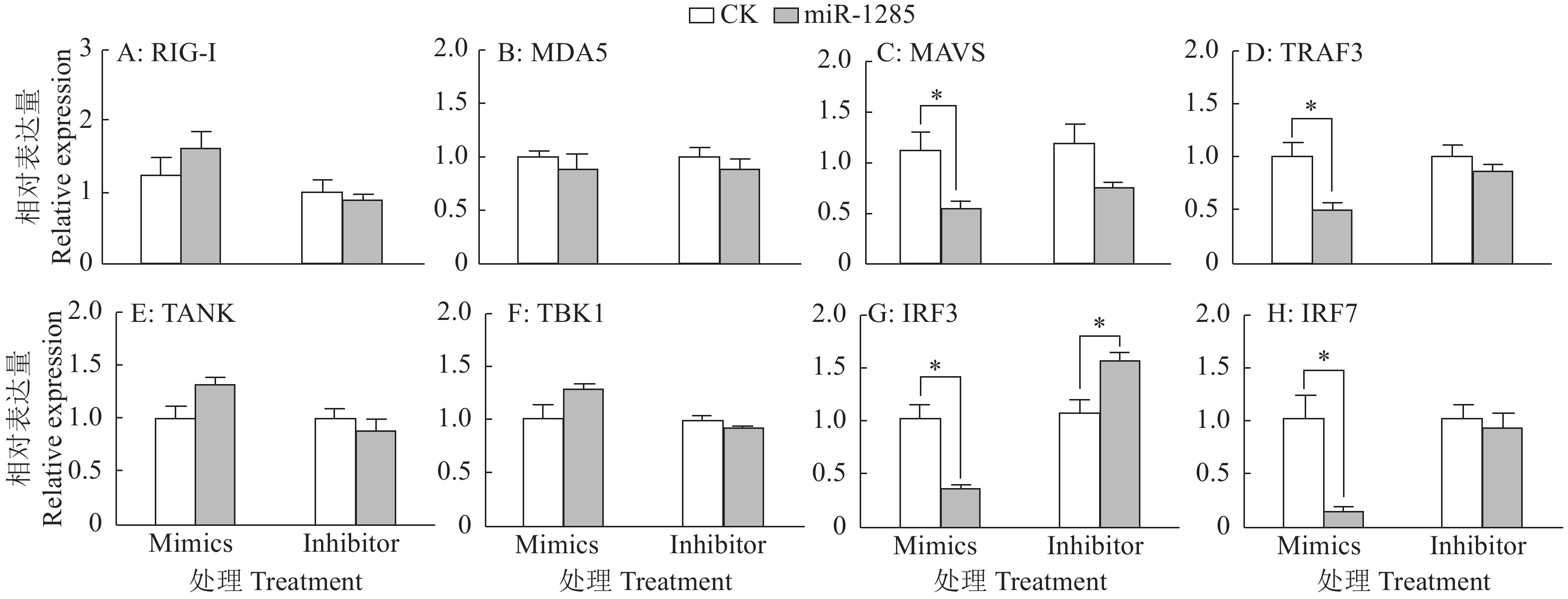
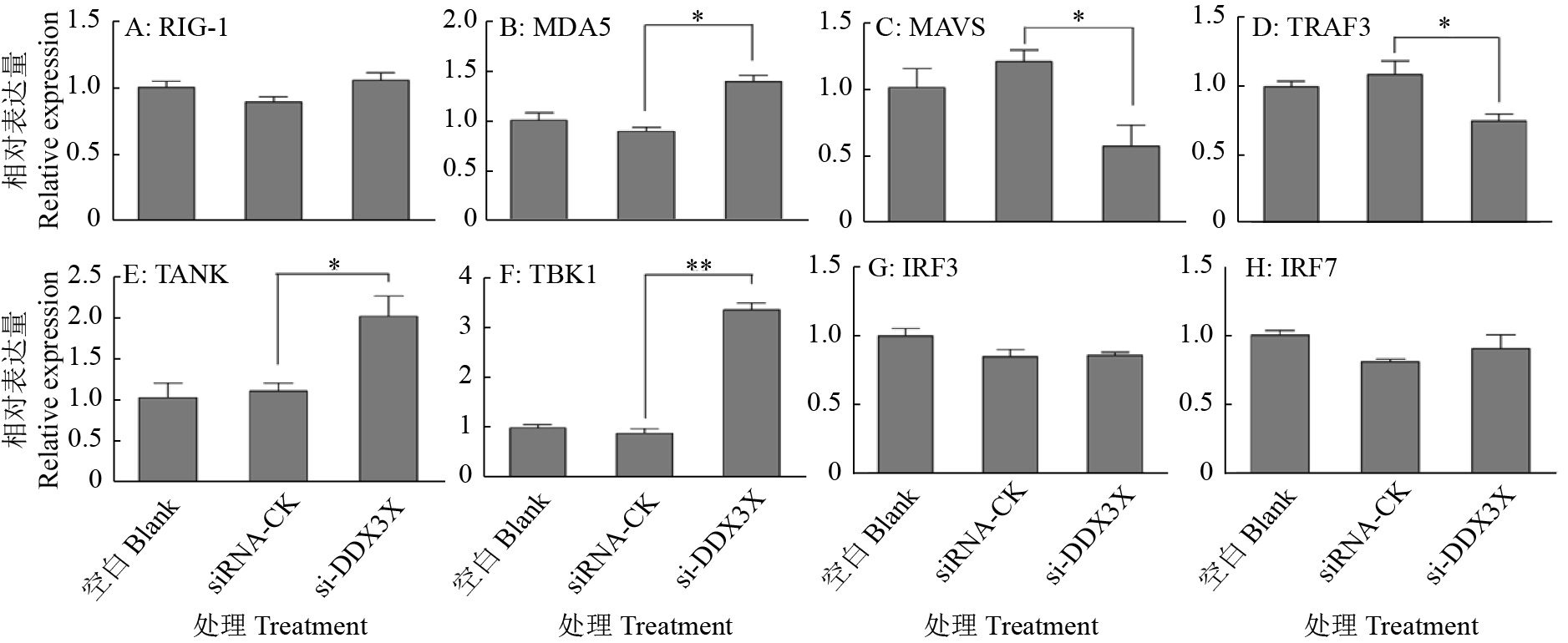
图 9 DDX3X沉默对RIG-I通路信号转导分子的影响
“*”和“**”分别表示处理与对照在P < 0.05和P < 0.01水平差异显著(Duncan’s法)
Figure 9. Effects of DDX3X silencing on signal transduction molecules of the RIG-I pathway
“*” and “**” represented statistical difference in comparison with control group at P < 0.05 and P < 0.01 levels respectively (Duncan’s method)
表 1 基因引物序列
Table 1 Primer sequences of genes
基因名称 Gene name 引物/探针序列(5′→3′) Primer/Probe sequence θ退火/ ℃ Annealing temperature 产物大小/bp Product size 文献 Reference RIG-I F: ATCCCAGCAACGAGAA 60 188 [36] R: GCCACGTCCAGTCAAT MDA5 F: GAGGAATCAGCACGAGGAA 58 73 [37] R: GTCAGTAATCCACTGGGA MAVS F: ATAGCCAGCCTTTCTCGG 60 237 [36] R: TAGCCTCAGTCTTGACCTCTTC TRAF3 F: GTGTCAAGAAGGCATCG 60 164 [36] R: CCTCAAACTGGCAATCA TANK F: GGACGCCTTGAACTACCTGT 60 119 R: GCCTGCCGAAAGGCTTCATA TBK1 F: GCCTTTCTCGGGGTCTTCAA 60 74 R: ACACTTTTCCTGATCCGCCT IRF3 F: CCAGTGGTGCCTACACTCCT 61 191 [38] R: AGAGGTGTCTGGCTCAGGAA IRF7 F: CGCCTCCTGGAAAACCAA 60 76 [37] R: CCCTGAGTTGTCCTGCAACA IFN-β F: GCTAACAAGTGCATCCTCCAAA 60 77 [39] R:AGCACATCATAGCTCATGGAAAGA GAPDH F: ACATGGCCTCCAAGGAGTAAGA 60 106 [40] R: GATCGAGTTGGGGCTGTGACT SVA-3C F: GAGCTTCAATCTCCTAGA 59 115 R: GTGTCATCATTCTCGTTAG 探针 Probe CAGACATTCGAGCCAAGCAACAA 69 -
[1] YATES L A, NORBURY C J, GIBERT R J C. The long and short of microRNA[J]. Cell, 2013, 153(3): 516-519. doi: 10.1016/j.cell.2013.04.003
[2] CHENG J, WU R, LONG L, et al. MiRNA-451a targets IFN regulatory factor 8 for the progression of systemic lupus erythematosus[J]. Inflammation, 2017, 40(2): 676-687. doi: 10.1007/s10753-017-0514-8
[3] WANG L, ZHOU L, HU D, et al. Porcine reproductive and respiratory syndrome virus suppresses post-transcriptionally the protein expression of IFN-β by upregulating cellular microRNAs in porcine alveolar macrophages in vitro[J]. Experimental and Therapeutic Medicine, 2018, 15(1): 115-126.
[4] MORIN R D, O’CONNOR M D, GRIFFITH M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells[J]. Genome Research, 2008, 18(4): 610-621. doi: 10.1101/gr.7179508
[5] ANTHON C, TAFER H, HAVGAARD J H, et al. Structured RNAs and synteny regions in the pig genome[J]. BMC Genomics, 2014, 15: 459. doi: 10.1186/1471-2164-15-459.
[6] GAO X, WANG Y, ZHAO H, et al. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma[J]. Oncotarget, 2016, 7(37): 59664-59675. doi: 10.18632/oncotarget.11198
[7] BORRELLI N, DENARO M, UGOLINI C, et al. MiRNA expression profiling of ‘noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas[J]. Modern Pathology, 2017, 30(1): 39-51. doi: 10.1038/modpathol.2016.157
[8] LIU J, YAN J, ZHOU C, et al. MiR-1285-3p acts as a potential tumor suppressor miRNA via down regulating JUN expression in hepatocellular carcinoma[J]. Tumor Biology, 2015, 36: 219-225. doi: 10.1007/s13277-014-2622-5
[9] HUANG H, XIONG G, SHEN P, et al. MicroRNA-1285 inhibits malignant biological behaviors of human pancreatic cancer cells by negative regulation of YAP1[J]. Neoplasma, 2017, 64(3): 358-366. doi: 10.4149/neo_2017_306
[10] LIAO J, LI Q, HU Z, et al. Mitochondrial miR-1285 regulates copper-induced mitochondrial dysfunction and mitophagy by impairing IDH2 in pig jejunal epithelial cells[J]. Journal of Hazardous Materials, 2022, 422: 126899. doi: 10.1016/j.jhazmat.2021.126899.
[11] VENKATARAMANNAN S, GADEK M, CALVIELLO L, et al. DDX3X and DDX3Y are redundant in protein synthesis[J]. RNA, 2021, 27(12): 1577-1588. doi: 10.1261/rna.078926.121
[12] VAN VOSS M R H, KAMMERS K, VESUNA F, et al. Global effects of DDX3 inhibition on cell cycle regulation identified by a combined phosphoproteomics and single cell tracking approach[J]. Translational Oncology, 2018, 11(3): 755-763. doi: 10.1016/j.tranon.2018.04.001
[13] HEATON S M, BORG N A, DIXIT V M. Ubiquitin in the activation and attenuation of innate antiviral immunity[J]. Journal of Experimental Medicine, 2016, 213(1): 1-13. doi: 10.1084/jem.20151531
[14] HATHAICHOTI S, VISITNONTHACHAI D, NGAMSIRI P, et al. Paraquat induces extrinsic pathway of apoptosis in A549 cells by induction of DR5 and repression of anti-apoptotic proteins, DDX3 and GSK3 expression[J]. Toxicology in Vitro, 2017, 42: 123-129. doi: 10.1016/j.tiv.2017.04.016
[15] LIN T C. DDX3X multifunctionally modulates tumor progression and serves as a prognostic indicator to predict cancer outcomes[J]. International Journal of Molecular Sciences, 2019, 21(1): 281. doi: 10.3390/ijms21010281.
[16] SOULAT D, BŰRCKSTŰMMER T, WESTERMAYER S, et al. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response[J]. EMBO Journal, 2008, 27(15): 2135-2146. doi: 10.1038/emboj.2008.126
[17] SCHRŐDER M, BARAN M, BOWIE A G. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation[J]. EMBO Journal, 2008, 27(15): 2147-2157. doi: 10.1038/emboj.2008.143
[18] OSHIUMI H, SAKAI K, MATSUMOT M, et al. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-β-inducing potential[J]. European Journal of Immunology, 2010, 40(4): 940-948. doi: 10.1002/eji.200940203
[19] SZAPPANOS D, TSCHISMAROV R, PERLOT T, et al. The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity[J]. PLoS Pathogens, 2018, 14(11): e1007397. doi: 10.1371/journal.ppat.1007397
[20] ADAMS M J, LEFKOWITZ E J, KING A M Q, et al. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses[J]. Archives of Virology, 2016, 161(10): 2921-2949. doi: 10.1007/s00705-016-2977-6
[21] VANNUCCI F A, LINHARES D C L, BARCELLOS D E S N, et al. Identification and complete genome of Seneca Valley virus in vesicular fluid and sera of pigs affected with idiopathic vesicular disease, Brazil[J]. Transboundary Emerging Disease, 2015, 62: 589-593. doi: 10.1111/tbed.12410
[22] CANNING P, CANON A, BATES J L, et al. Neonatal mortality, vesicular lesions and lameness associated with Senecavirus A in a U. S. sow farm[J]. Transboundary and Emerging Disease, 2016, 63(4): 373-378. doi: 10.1111/tbed.12516
[23] ZHANG X L, ZHU Z X, YANG F, et al. Review of Seneca Valley virus: A call for increased surveillance and research[J]. Frontiers in Microbiology, 2018, 9: 940. doi: 10.3389/fmicb.2018.00940.
[24] FERNANDES M H V, MAGGIOLI M F, OTTA J, et al. Senecavirus A 3C protease mediates host cell apoptosis late in infection[J]. Frontiers in Immunology, 2019, 10: 363. doi: 10.3389/fimmu.2019.00363.
[25] LEME R A, OLIVEIRA T E S, ALFIERI A F, et al. Pathological, immunohistochemical and molecular findings associated with Senecavirus A-induced lesions in neonatal piglets[J]. Journal of Comparative Pathology, 2016, 155(2/3): 145-155.
[26] HAUSE B M, MYERS O, DUFF J, et al. Senecavirus A in pigs, United States, 2015[J]. Emerging Infectious Diseases, 2016, 22(7): 1323-1325. doi: 10.3201/eid2207.151591
[27] XU W, HOLE K, GOOLIA M, et al. Genome wide analysis of the evolution of Senecavirus A from swine clinical material and assembly yard environmental samples[J]. PLoS One, 2017, 12(5): e0176964. doi: 10.1371/journal.pone.0176964
[28] WU Q, ZHAO X, BAI Y, et al. The first identification and complete genome of Senecavirus A affecting pig with idiopathic vesicular disease in China[J]. Transboundary and Emerging Disease, 2017, 64(5): 1633-1640. doi: 10.1111/tbed.12557
[29] SAENG-CLUTO K, RODTIAN P, TEMEEYASEN G, et al. The first detection of Senecavirus A in pigs in Thailand, 2016[J]. Transboundary and Emerging Disease, 2018, 65(1): 285-288. doi: 10.1111/tbed.12654
[30] SUN D, VANNUCCI F, KNUTSON T P, et al. Emergence and whole-genome sequence of Senecavirus A in Colombia[J]. Transboundary and Emerging Diseases, 2017, 64(5): 1346-1349. doi: 10.1111/tbed.12669
[31] ARZT J, BERTRAM M R, VU L T, et al. First detection and genome sequence of Senecavirus A in Vietnam[J]. Microbiology Resource Announcements, 2019, 8(3). doi: 10.1128/MRA.01247-18.
[32] SUN Y, CHENG J, WU R T, et al. Phylogenetic and genome analysis of 17 novel Senecavirus A isolates in Guangdong Province, 2017[J]. Frontiers in Veterinary Science, 2018, 5: 314. doi: 10.3389/fvets.2018.00314.
[33] QIAN S, FAN W, QIAN P, et al. Isolation and full-genome sequencing of Seneca Valley virus in piglets from China, 2016[J]. Virology Journal, 2016, 13: 173. doi: 10.1186/s12985-016-0631-2.
[34] ZHU Z, YANG F, CHEN P, et al. Emergence of novel Seneca Valley virus strains in China, 2017[J]. Transboundary and Emerging Diseases, 2017, 64(4): 1024-1029. doi: 10.1111/tbed.12662
[35] WANG H, LI C, ZHAO B, et al. Complete genome sequence and phylogenetic analysis of Senecavirus A isolated in Northeast China in 2016[J]. Archives of Virology, 2017, 162(10): 3173-3176. doi: 10.1007/s00705-017-3480-4
[36] ZHANG J, MIAO J, HOU J, et al. The effects of H3N2 swine influenza virus infection on TLRs and RLRs signaling pathways in porcine alveolar macrophages[J]. Virology Journal, 2015, 12: 61. doi: 10.1186/s12985-015-0284-6.
[37] 谢立兰. 伪狂犬病毒和猪传染性胃肠炎病毒诱导β干扰素产生的分子机制研究[D]. 武汉: 华中农业大学, 2011. [38] ISLAM M A, GROßE-BRINKHAUS C, PRŐLL M J, et al. Deciphering transcriptome profiles of peripheral blood mononuclear cells in response to PRRSV vaccination in pigs[J]. BMC Genomics, 2016, 17: 641. doi: 10.1186/s12864-016-2849-1.
[39] 王荡. 口蹄疫病毒Lpro和3Cpro调控宿主抗病毒天然免疫反应的分子机制研究[D]. 武汉: 华中农业大学, 2011. [40] 王国庆. 口蹄疫病毒2B蛋白拮抗RIG-I抗病毒作用研究[D]. 兰州: 甘肃农业大学, 2015. [41] DERRICK T, ROBERTS C H, RAJASEKHAR M, et al. Conjunctival microRNA expression in inflammatory trachomatous scarring[J]. PLoS Neglected Tropical Diseases, 2013, 7(3): e2117. doi: 10.1371/journal.pntd.0002117
[42] KAUR S, KRISHN S R, RACHAGANI S, et al. Significance of microRNA-based biomarkers for pancreatic cancer[J]. Annals of Translational Medicine, 2015, 3(18): 277.
[43] LIU Y, XU X, XU X, et al. MicroRNA-193a-3p inhibits cell proliferation in prostate cancer by targeting cyclin D1[J]. Oncology Letters, 2017, 14(5): 5121-5128.
[44] MORGUL M H, KLUNK S, ANASTASIADOU Z, et al. Diagnosis of HCC for patients with cirrhosis using miRNA profiles of the tumor-surrounding tissue: A statistical model based on stepwise penalized logistic regression[J]. Experimental and Molecular Pathology, 2016, 101(2): 165-171. doi: 10.1016/j.yexmp.2016.07.014
[45] POIRIER J T, DOBROMILSKAVA I, MORIARTY W F, et al. Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer[J]. Journal of the National Cancer Institute, 2013, 105(14): 1059-1065. doi: 10.1093/jnci/djt130
[46] ZHOU Z H, SUN Y, YAN X L, et al. Swine acute diarrhea syndrome coronavirus (SADS-CoV) antagonizes interferon-β production via blocking IPS-1 and RIG-I[J]. Virus Research, 2020, 278: 197843. doi: 10.1016/j.virusres.2019.197843.
[47] ANDREJEVA J, CHILDS K S, YOUNG D F, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(49): 17264-17269. doi: 10.1073/pnas.0407639101
[48] CAO L, GE X, GAO Y, et al. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-beta production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway[J]. Virology Journal, 2015, 12: 127. doi: 10.1186/s12985-015-0345-x.
[49] CHEN Q, LIU Q, LIU D, et al. Molecular cloning, functional characterization and antiviral activity of porcine DDX3X[J]. Biochemical and Biophysical Research Communications, 2014, 443(4): 1169-1175. doi: 10.1016/j.bbrc.2013.12.098
[50] ZHAO X, WU Q, BAI Y, et al. Phylogenetic and genome analysis of seven Senecavirus A isolates in China[J]. Transboundary and Emerging Diseases, 2017, 64(6): 2075-2082. doi: 10.1111/tbed.12619
[51] THOMPSON S R, SARNOW P. Regulation of host cell translation by viruses and effects on cell function[J]. Current Opinion in Microbiology, 2000, 3(4): 366-370. doi: 10.1016/S1369-5274(00)00106-5
[52] QIAN S, FAN W, LIU T, et al. Seneca Valley virus suppresses host type I interferon production by targeting adaptor proteins MAVS, TRIF, and TANK for cleavage[J]. Journal of Virology, 2017, 91(16): e00823-17.
[53] LIU T, LI X, WU M, et al. Seneca Valley virus 2C and 3Cpro induce apoptosis via mitochondrion-mediated intrinsic pathway[J]. Frontiers in Microbiology, 2019, 10: 1202. doi: 10.3389/fmicb.2019.01202.
[54] XUE Q, LIU H, ZHU Z, et al. Seneca Valley Virus 3C protease negatively regulates the type I interferon pathway by acting as a viral deubiquitinase[J]. Antiviral Research, 2018, 160: 183-189. doi: 10.1016/j.antiviral.2018.10.028
-
期刊类型引用(0)
其他类型引用(1)





 下载:
下载: