Identification and expression of SR45 subfamily gene in Manihot esculenta
-
摘要:目的
精氨酸和丝氨酸富集蛋白(Arginine/serine-rich proteins,SR)为剪接复合体的主要成员,不仅参与植物前体mRNA可变剪接过程,在植物非生物胁迫中也具有相当重要的作用。SR45基因为SR基因亚家族成员之一。本研究旨在分析木薯SR45亚家族成员蛋白结构特征与表达模式,为进一步了解该亚家族基因在木薯中的功能提供理论支持。
方法利用生物信息学技术重新构建木薯SR基因家族进化树,对木薯SR45亚家族蛋白理化性质、基因结构、保守结构域进行分析,同时利用转录组数据分析低温与干旱胁迫下SR基因表达变化,运用实时荧光定量PCR(RT-qPCR)研究各基因成员在不同组织中的特异性表达以及对低温胁迫的响应。
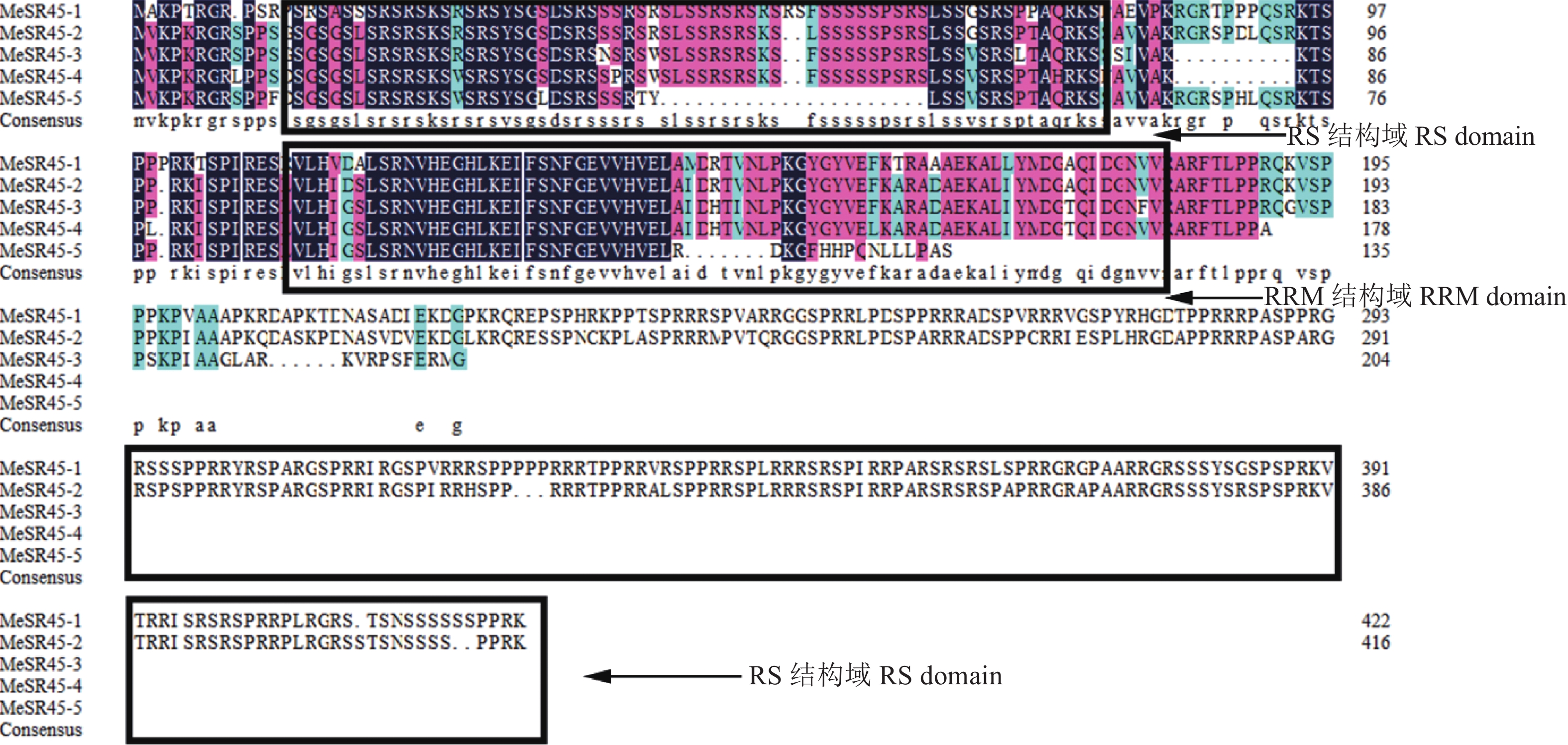
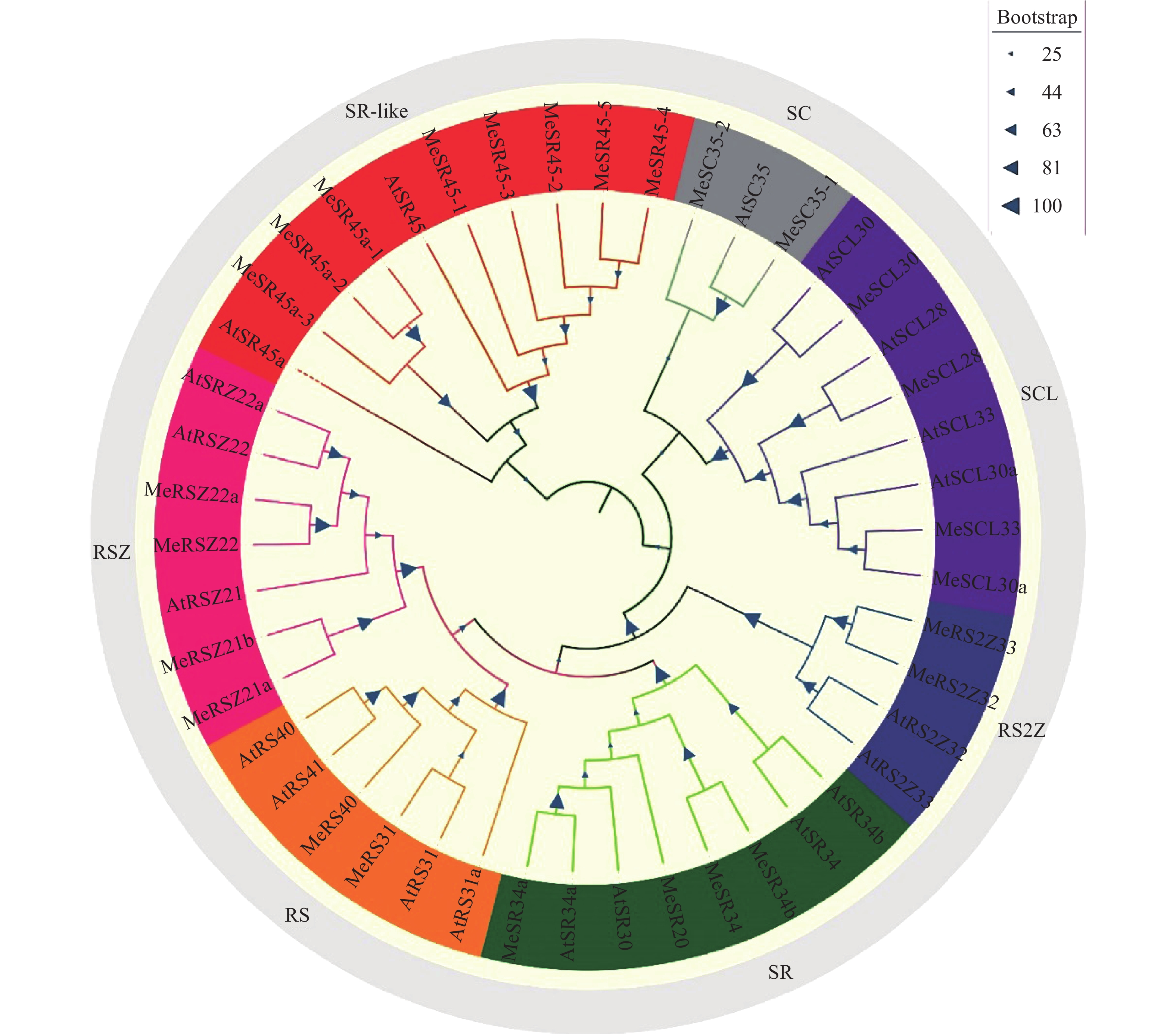
结果木薯SR基因家族共7个大类26个成员,SR45亚家族共有5个成员;SR45亚家族基因编码蛋白长度为135~423 aa,相对分子质量为14970~47210,等电点为5.19~12.34;预测其主要定位于细胞核和叶绿体;SR45编码RS结构域和RRM结构域,具有Motif 3、Motif 6、Motif 2和Motif 1保守基序。转录组数据和RT-qPCR分析表明,SR45基因均响应木薯低温胁迫,且MeSR45-2显著上调表达,MeSR45-4、MeSR45-5在木薯根和叶中有较高表达量。
结论木薯SR45亚家族基因显著响应低温胁迫;将MeSR45-2基因列为调控低温逆境变化的候选基因,将根和叶列为研究SR45基因亚类成员的主要组织。本研究为探索木薯SR45基因奠定了理论基础,并为进一步研究木薯SR45基因逆境胁迫应答过程指明了方向。
Abstract:ObjectiveAs major members of the splicing complex, arginine/serine-rich proteins (SR) not only participate in the process of alternative splicing of plant precursor mRNA, but also play an important role in abiotic stress. SR45 gene is a member of SR gene subfamily, this study was aimed to analyze the structural characteristics and expression patterns of SR45 subfamily proteins in cassava, and provide a theoretical support for further understanding the functions of SR45 subfamily genes in cassava.
MethodThe evolutionary tree of cassava SR gene family was reconstructed by bioinformatics, and the physical and chemical properties, gene structure and conserved domain of cassava SR45 subfamily protein were analyzed. At the same time, transcriptome data were used to analyze the changes of SR gene expression under low temperature and drought stress, quantitative real-time PCR (RT-qPCR) was used to study the specific expression of SR gene members in different tissues and their responses to low temperature stress.
ResultThe cassava SR gene family consisted of 26 members in seven categories, and the SR45 subfamily consisted of five members. The length of proteins encoded by SR45 subfamily genes were 135–417 aa, with relative molecular mass being 14970–47210, PI being 5.19–12.34. It was predicted that they were mainly located in the nucleus and chloroplast. The RS domain and RRM domain encoded by SR45 had Motif 3, Motif 6, Motif 2 and Motif 1 conserved motifs. Transcriptional data and RT-qPCR analysis showed that all SR45 genes were responsive to low temperature stress in cassava, MeSR45-2 was significantly up-regulated, MeSR45-4and MeSR45-5 were highly expressed in roots and leaves of cassava.
ConclusionSR45 subfamily genes of cassava significantly respond to low temperature stress. MESR45-2 gene is selected as the candidate gene for regulating low temperature stress, root and leaf are the main tissues for SR45 subfamily gene research. This study lays a theoretical foundation for the exploration of cassava SR45 gene, and points out a direction for further research on cassava SR45 gene responsive to stress.
-
Keywords:
- Cassava /
- Alternative splicing /
- SR gene family /
- SR45 gene /
- Low temperature stress
-
-
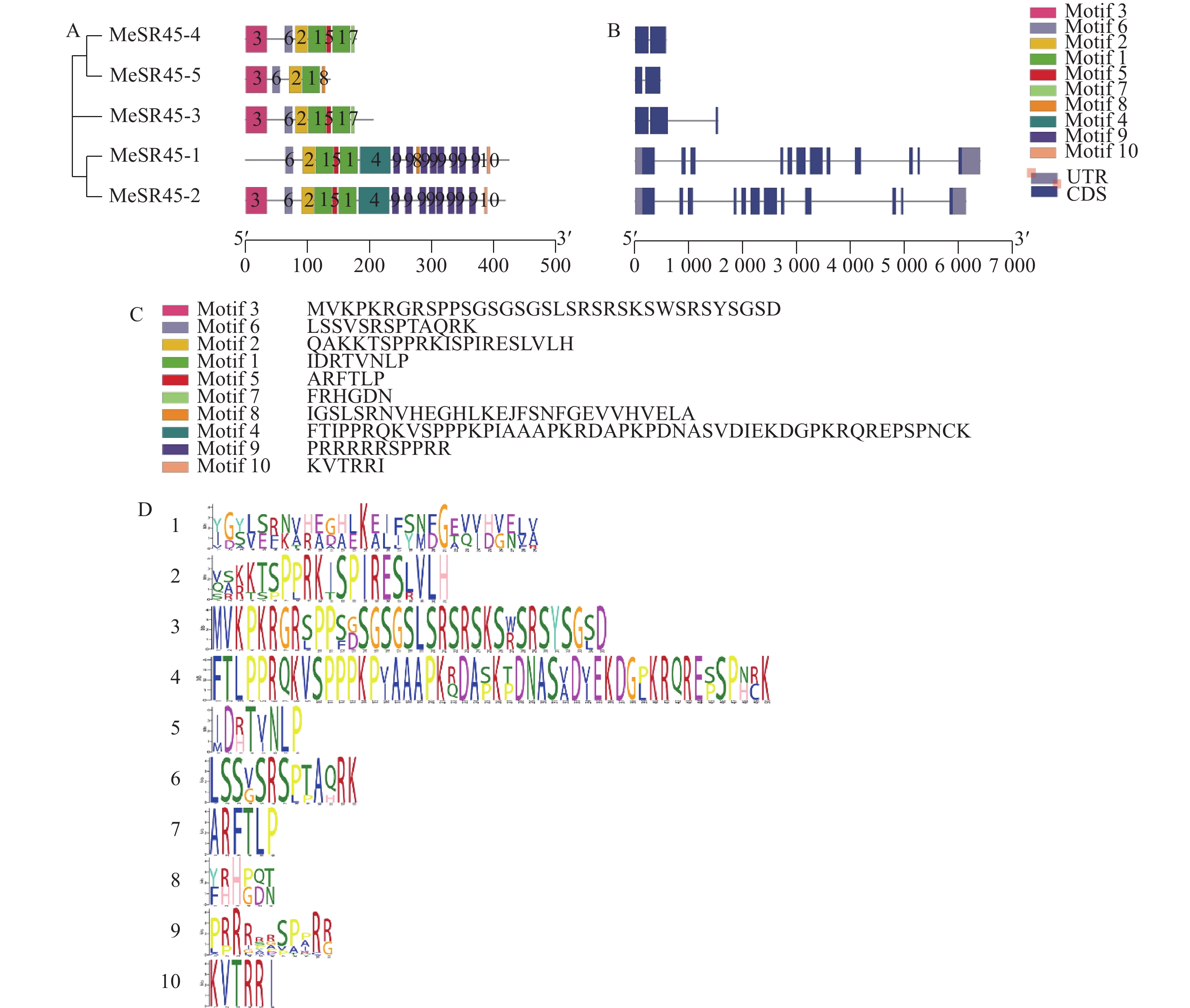
图 2 MeSR45蛋白保守Motif结构和基因结构
A:MeSR45基因家族隐马尔可夫模型图案的位置;B:MeSR45基因结构;C:MeSR45蛋白保守Motif序列;D:MeSR45基因家族隐马尔可夫模型图
Figure 2. Consevered motif and gene structures of MeSR45 protein
A: Location of Hidden Markov model of MeSR45 gene family; B: MeSR45 gene structure; C: Conserved motif sequence of MeSR45; D: The map of Hidden Markov model of MeSR45 gene family
图 5 低温胁迫下不同处理时间木薯MeSR45基因表达模式
“*”“**”“***”分别表示相关基因在不同处理时间与对照(0 h)在P < 0.05、P < 0.01和P < 0.001水平差异显著(t检验)
Figure 5. MeSR45 gene expression pattern in cassava under low temperature stress at different treatment time
“*” “**” and “***” indicate significant differences of the related genes between the treatment time and control (0 h) at the levels of P < 0.05, P < 0.01 and P < 0.001, respectively (t test)
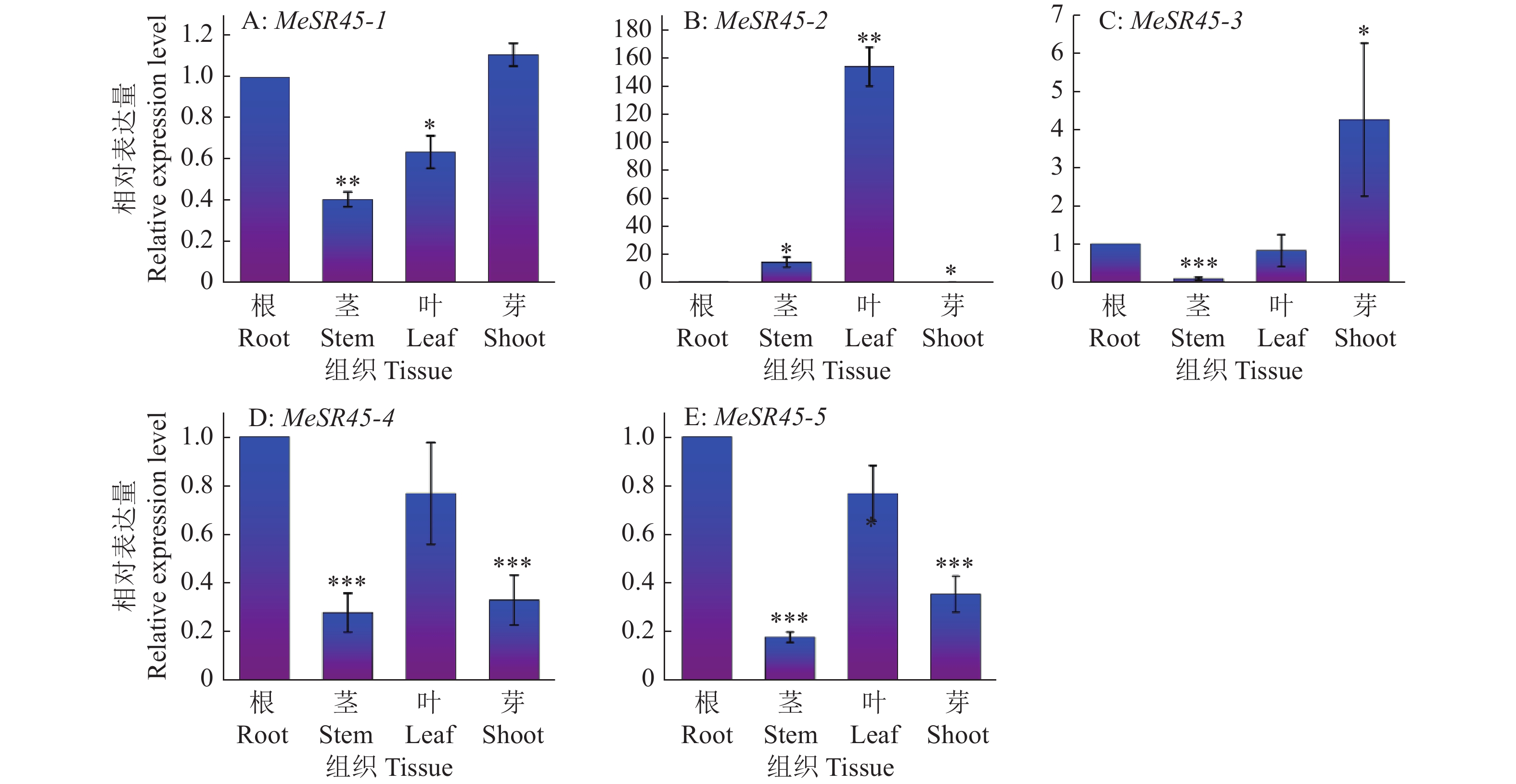
图 6 MeSR45亚家族基因在木薯不同组织部位中的表达模式
“*”“**”“***”分别表示相关基因在处理组织部位与对照(根)在P<0.05、P<0.01和P<0.001水平差异显著(t检验)
Figure 6. Expression patterns of MeSR45 subfamily gene in different cassava tissues
“*” “**” and “***” indicate significant differences of the related genes between the treatment tissue and control (root) at the levels of P < 0.05, P < 0.01 and P<0.001, respectively (t test)
表 1 MeSR45基因RT-qPCR引物序列
Table 1 Primer sequences of MeSR45 genes for RT-qPCR
基因名称
Gene name上游引物(5′→3′)
Forward primer (5′→3′)下游引物(5′→3′)
Reverse primer (5′→3′)MeActin TGATGAGTCTGGTCCATCCA CCTCCTACGACCCAATCTCA MeCOR ACACGACAGGATCTGCCAAC GGCCAGATGGAACATGAGCA MeSR45-1 GTCACGCTCTTACTCCGGTT TGAAGACGAAGAGAACGATCGAG MeSR45-2 CTCCGCGACAGAAGGTTTCA GTCGCCTTGGAGGAGAATGT MeSR45-3 GGATGGGACCCAGATTGATGG GCCTGACTTTACGAGCGAGG MeSR45-4 TTCTGGCTCGGGTTCATTGT AGCAGTAGGGCTGCGACTAA MeSR45-5 TACCTCAGCTCTGTTAGCCG GAGAACGACCTCGTTTAGCCA -
[1] COCK J H. Cassava: A basic energy source in the tropics[J]. Science, 1982, 218(4574): 755-762. doi: 10.1126/science.7134971
[2] HUANG L, YE Z, BELL R W, et al. Boron nutrition and chilling tolerance of warm climate crop species[J]. Annals of Botany, 2005, 96(5): 755-767. doi: 10.1093/aob/mci228
[3] LEVIATAN N, ALKAN N, LESHKOWITZ D, et al. Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray[J]. PLoS One, 2013, 8(6): e66511. doi: 10.1371/journal.pone.0066511
[4] SHEN Y F, WU X P, LIU D M, et al. Cold-dependent alternative splicing of a Jumonji C domain-containing gene MtJMJC5 in Medicago truncatula[J]. Biochemical and Biophysical Research Communications, 2016, 474(2): 271-276. doi: 10.1016/j.bbrc.2016.04.062
[5] IIDA K, SEKI M, SAKURAI T, et al. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences[J]. Nucleic Acids Research, 2004, 32(17): 5096-5103. doi: 10.1093/nar/gkh845
[6] THATCHER S R, DANILEVSKAYA O N, MENG X, et al. Genome-wide analysis of alternative splicing during development and drought stress in maize[J]. Plant Physiology, 2016, 170(1): 586-599. doi: 10.1104/pp.15.01267
[7] LIU Z S, QIN J X, TIAN X J, et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.)[J]. Plant Biotechnology Journal, 2018, 16(3): 714-726. doi: 10.1111/pbi.12822
[8] REDDY A S N, ALI G S. Plant serine/arginine-rich proteins: Roles in precursor messenger RNA splicing, plant development, and stress responses[J]. Wiley Interdisciplinary Reviews: RNA, 2011, 2(6): 875-889. doi: 10.1002/wrna.98
[9] LI S X, YU X, CHENG Z H, et al. Large-scale analysis of the cassava transcriptome reveals the impact of cold stress on alternative splicing[J]. Journal of Experimental Botany, 2020, 71(1): 422-434.
[10] LI Y, GUO Q, LIU P, et al. Dual roles of the serine/arginine-rich splicing factor SR45a in promoting and interacting with nuclear cap-binding complex to modulate the salt-stress response in Arabidopsis[J]. New Phytologist, 2021, 230(2): 641-655. doi: 10.1111/nph.17175
[11] PALUSA S G, ALI G S, REDDY A S N. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses[J]. Plant Journal, 2007, 49(6): 1091-1107. doi: 10.1111/j.1365-313X.2006.03020.x
[12] CARVALHO R F, CARVALHO S D, DUQUE P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis[J]. Plant Physiology, 2010, 154(2): 772-783. doi: 10.1104/pp.110.155523
[13] PANDIT S, ZHOU Y, SHIUE L, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing[J]. Molecular Cell, 2013, 50(2): 223-235. doi: 10.1016/j.molcel.2013.03.001
[14] BARTA A, KALYNA M, LORKOVIC Z J. Plant SR proteins and their functions[J]. Nuclear Pre-mRNA Processing in Plants, 2008, 326: 83-102.
[15] BARTA A, KALYNA M, REDDY A S. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants[J]. Plant Cell, 2010, 22(9): 2926-2929. doi: 10.1105/tpc.110.078352
[16] CHEN S, LI J, LIU Y, et al. Genome-wide analysis of serine/arginine-rich protein family in wheat and Brachypodium distachyon[J]. Plants, 2019, 8(7): 188. doi: 10.3390/plants8070188.
[17] KALYNA M, BARTA A. A plethora of plant serine/arginine-rich proteins: Redundancy or evolution of novel gene functions?[J]. Biochemical Society Transactions, 2004, 32: 561-564. doi: 10.1042/BST0320561
[18] GU J, MA S, ZHANG Y, et al. Genome-wide identification of cassava serine/arginine-rich proteins: Insights into alternative splicing of pre-mRNAs and response to abiotic stress[J]. Plant and Cell Physiology, 2020, 61(1): 178-191. doi: 10.1093/pcp/pcz190
[19] 马思雅. 木薯SR蛋白家族的系统命名及表达分析[D]. 海口: 海南大学, 2018. [20] ZHAO C, ZAYED O, ZENG F, et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis[J]. New Phytologist, 2019, 224(1): 274-290. doi: 10.1111/nph.15867
[21] 顾进宝. 编码木薯SR蛋白基因的选择性剪接及其调控拟南芥盐胁迫响应中的作用研究[D]. 合肥: 合肥工业大学, 2020. [22] ALBAQAMI M, LALUK K, REDDY A S N. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner[J]. Plant Molecular Biology, 2019, 100(4/5): 379-390.
[23] CRUZ T M D, CARVALHO R F, RICHARDSON D N, et al. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression[J]. International Journal of Molecular Sciences, 2014, 15(10): 17541-17564. doi: 10.3390/ijms151017541
[24] DAY I S, GOLOVKIN M, PALUSA S G, et al. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing[J]. Plant Journal, 2012, 71(6): 936-947. doi: 10.1111/j.1365-313X.2012.05042.x
[25] ALI G S, PALUSA S G, GOLOVKIN M, et al. Regulation of plant developmental processes by a novel splicing factor[J]. PLoS One, 2007, 2(5): e471. doi: 10.1371/journal.pone.0000471
[26] ZHANG X N, MOUNT S M. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development[J]. Plant Physiology, 2009, 150(3): 1450-1458. doi: 10.1104/pp.109.138180
[27] PARK H J, YOU Y N, LEE A, et al. OsFKBP20-1b interacts with the splicing factor OsSR45 and participates in the environmental stress response at the post-transcriptional level in rice[J]. Plant Journal, 2020, 102(5): 992-1007. doi: 10.1111/tpj.14682
[28] TANABE N, KIMURA A, YOSHIMURA K, et al. Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus[J]. Plant Molecular Biology, 2009, 70(3): 241-252. doi: 10.1007/s11103-009-9469-y
[29] XING D, WANG Y, HAMILTON M, et al. Transcriptome-wide identification of RNA targets of Arabidopsis serine/arginine-rich45 uncovers the unexpected roles of this RNA binding protein in RNA processing[J]. Plant cell, 2015, 27(12): 3294-3308. doi: 10.1105/tpc.15.00641
[30] XIAO J, HU R, GU T, et al. Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat[J]. BMC Genomics, 2019, 20: 287. doi: 10.1186/s12864-019-5632-2.



 下载:
下载: