Analyses of Ipomoea batatas cultivated species and wild relatives based on mtDNA and cpDNA sequences
-
摘要:目的
利用线粒体DNA(Mitochondrial DNA,mtDNA)序列和叶绿体DNA(Chloroplast DNA,cpDNA)matK序列对甘薯Ipomoea batatas栽培种及近缘野生种进行分子鉴定和亲缘关系分析,为甘薯栽培种和近缘野生种的种质鉴定、保护及开发利用提供理论依据。
方法以3个甘薯栽培种及8个近缘野生种为材料,采用CTAB法提取基因组DNA,通过PCR扩增mtDNA序列和cpDNA matK序列,使用DnaSP 6.0对序列进行核苷酸多态性、单倍型多样性等特征分析,并基于邻接法构建3个甘薯栽培种及8个近缘野生种的系统发育进化树。
结果5个mtDNA序列和cpDNA matK序列经测序、比对、拼接后,长度为6 713 bp,GC占比在47.79%~48.31%之间。合并序列的单倍型数量、核苷酸多态性、变异位点数量、单一突变位点数量、简约信息位点数量和插入/缺失位点数量分别为9、0.003 25、69、39、30和111。中性检验显示,合并序列差异不显著(P>0.10),遵循中性进化模型。3个甘薯栽培种及8个近缘野生种间的遗传距离在0.000 00~0.005 84之间,平均遗传距离0.003 26,遗传多样性较低;按照亲缘关系被分为2大类,各大类内部亲缘关系较近。
结论本研究采用的序列可对甘薯栽培种和近缘野生种进行准确的鉴定区分,为甘薯近缘野生种的进化和利用提供参考和理论指导。
Abstract:ObjectiveTo conduct molecular identification and genetic relationship analysis of Ipomoea batatas cultivated species and wild relatives based on mitochondrial DNA (mtDNA) and chloroplast DNA (cpDNA) matK sequences, and provide theoretical bases for germplasm identification, protection, development and utilization.
MethodThree cultivated species and eight wild relatives were used as materials, from which total DNA was extracted by the CTAB method. Their mtDNA and cpDNA matK sequences were amplified by PCR. DnaSP 6.0 was used to analyze nucleotide diversity, haplotype diversity and other characteristics. The phylogenetic tree of three cultivated species and eight wild relatives was constructed based on the neighbor-joining method.
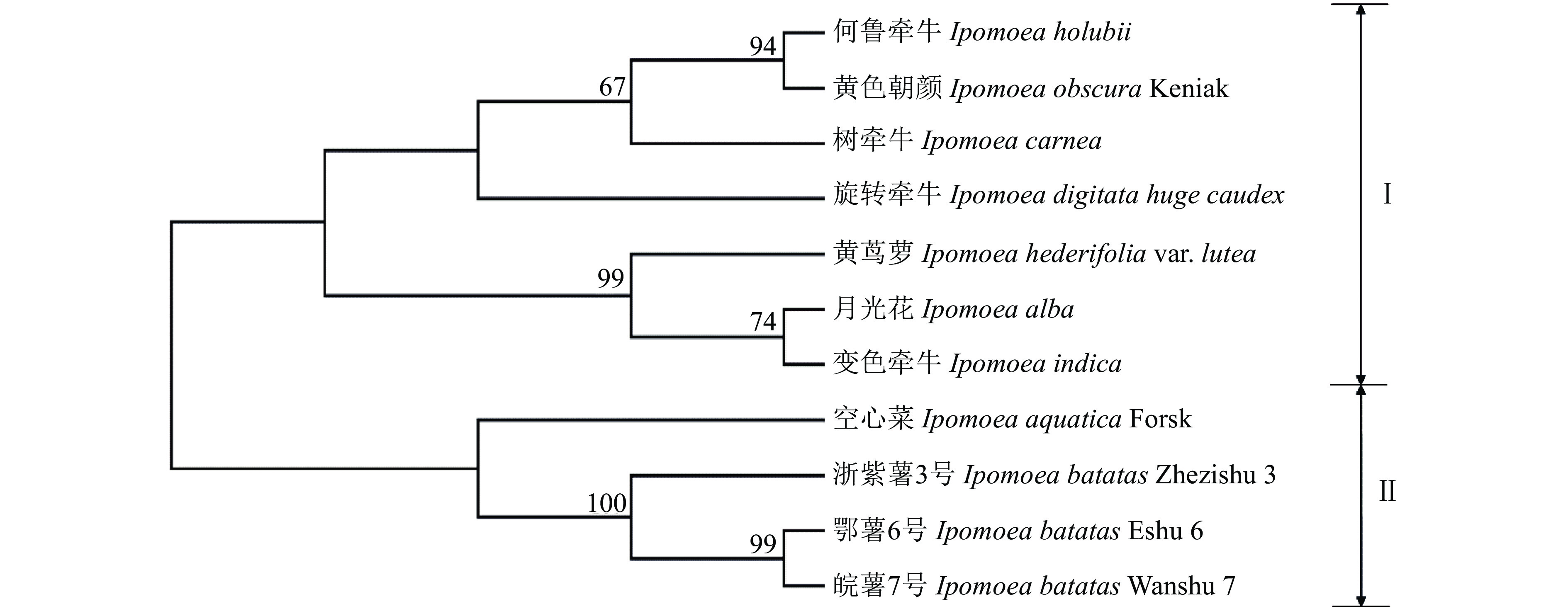
ResultThe length of five mtDNA regions and one cpDNA region was 6 713 bp after sequencing, alignment and splicing, the GC proportion was 47.79%−48.31%, and the haplotype number, nucleotide diversity, variable site number, singleton variable site number, parsimony informative site number, insertion/deletion site number were 9, 0.003 25, 69, 39, 30, 111, respectively. The neutrality test showed there was no significant difference between Tajima’sD values at the level of P>0.10, which indicated that variation of those regions followed neutral theory of molecular evolution. The genetic distances among three cultivated species and eight wild relatives ranged from 0.000 00 to 0.005 84, with an average genetic distance of 0.003 26, indicating low genetic diversity. The phylogenetic tree divided the 11 species into two categories with close genetic relationship within the category.
ConclusionThe sequences used in this study can accurately identify I. batatas cultivated species and wild relatives, and provide references and theoretical guidance for the evolution and utilization of I. batatas wild relatives.
-
Keywords:
- Ipomoea batatas /
- mitochondrial DNA /
- matK sequence /
- phylogeny /
- genetic diversity
-
-
表 1 试验采用引物信息
Table 1 The information of primers used in this experiment
类别
Species基因序列
Gene sequence引物
Primer片段大小/bp
Fragment size退火温度/℃
Annealing temperature参考文献
ReferencemtDNA ccb203 F: ASGTTCTACGGACCGATGCC 500 57 [22] R: CACGGGGAGGGAGCRGGCGA ccb256 F: GGAAGTTAGCAAAGTTAGAC 520 57 [22] R: TTGTTCTTAACAGCGATGGC nad2/1-2 F: TTTTCTTCCTCATTCTKATTT 1200 57 [22] R: CCACTCTATTGTCCACTTCTA nad2/4-5 F: TTCATATAGAATCCATGTCC 1800 57 [22] R: CTATTTGTTCTTCGCCGCTT nad5/4-5 F: CCAATTTTTGGGCCAATTCC 1400 57 [23] R: CATTGCAAAGGCATAATGAT nad7/1-2 F: ACCTCAACATCCTGCTGCTC 1200 57 [23] R: CGATCAGAATAAGGTAAAGC cpDNA matK F: CGTACAGTACTTTTGTGTTTACGAG 1500 57 [24] R: ACCCAGTCCATCTGGAAATCTTGGTTC 表 2 试验材料的序列多态性信息
Table 2 Sequence polymorphism information of experiment materials
基因序列
Gene
sequence长度/
bp
LengthGC占
比/%
GC
proportion核苷酸多态性
Nucleotide
diversity变异位点数
Variable
site
number单一突变位点数
Singleton
variable
site number简约信息位点数
Parsimony
informative
site number插入/缺失位点数
Insertion/
deletion
site numberccb203 494 46.52~47.17 0.003 20 5 2 3 6 nad2/1-2 1 247 53.90~54.69 0.000 17 1 1 0 11 nad2/4-5 1 503 48.02~48.54 0.003 43 12 3 9 63 nad5/4-5 1 665 45.93~46.50 0.003 95 20 10 10 6 nad7/1-2 964 56.39~57.23 0.001 86 4 0 4 24 matK 840 32.90~34.09 0.007 11 27 23 4 1 合并 Total 6 713 47.79~48.31 0.003 25 69 39 30 111 表 3 试验材料的单倍型多样性
Table 3 Haplotype diversity of experiment materials
基因序列
Gene
sequence单倍型数量
Haplotype
number单倍型多样性
Haplotype
diversity单倍型多样性方差
Variance of
haplotype diversity单倍型多样性标准差
Standard deviation of
haplotype diversityccb203 5 0.818 0.006 82 0.083 nad2/1-2 2 0.182 0.020 61 0.144 nad2/4-5 8 0.927 0.004 42 0.066 nad5/4-5 8 0.927 0.004 42 0.066 nad7/1-2 5 0.818 0.006 82 0.083 matK 9 0.945 0.004 34 0.066 合并 Total 9 0.945 0.004 34 0.066 表 4 Tajima’s D测验和Fu and Li’s D*/F*测验
Table 4 Tajima’s D test and Fu and Li’s D*/F* test
基因序列 Gene sequence Tajima’s D Fu and Li’s D* Fu and Li’s F* P ccb203 −0.321 97 −0.083 18 −0.161 10 > 0.10 nad2/1-2 −1.128 50 −1.289 46 −1.399 19 > 0.10 nad2/4-5 0.500 00 0.574 40 0.628 71 > 0.10 nad5/4-5 −0.356 59 −0.514 26 −0.537 17 > 0.10 nad7/1-2 1.018 28 1.214 66 1.312 69 > 0.10 matK −1.627 84 −2.009 96 −2.168 40 > 0.05, < 0.10 合并Total −0.639 26 −0.796 77 −0.858 74 > 0.10 表 5 11份试验材料的遗传距离1)
Table 5 Genetic distance among 11 experimental materials
样品编号
Sample No.1 2 3 4 5 6 7 8 9 10 1 2 0.003 47 3 0.002 99 0.003 63 4 0.003 78 0.001 42 0.004 42 5 0.002 99 0.005 21 0.004 73 0.005 84 6 0.003 15 0.001 10 0.003 78 0.000 94 0.004 89 7 0.002 52 0.003 15 0.003 63 0.003 78 0.004 26 0.003 15 8 0.001 89 0.003 15 0.002 68 0.004 10 0.003 63 0.003 47 0.002 52 9 0.003 47 0.003 63 0.003 15 0.003 78 0.004 89 0.003 15 0.003 15 0.003 15 10 0.003 47 0.003 63 0.003 15 0.003 78 0.004 89 0.003 15 0.003 15 0.003 15 0.000 00 11 0.003 47 0.003 63 0.003 15 0.003 78 0.004 89 0.003 15 0.003 15 0.003 15 0.000 00 0.000 00 1) 1:何鲁牵牛,2:‘黄茑萝’,3:空心菜,4:‘月光花’,5:‘黄色朝颜’,6:变色牵牛,7:旋转牵牛,8:树牵牛,9:‘浙紫薯3号’,10:‘鄂薯6号’,11:‘皖薯7号’
1) 1: Ipomoea holubii, 2: Ipomoea hederifolia var. lutea, 3: Ipomoea aquatica Forsk, 4: Ipomoea alba, 5: Ipomoea obscura Keniak, 6: Ipomoea indica, 7: Ipomoea digitata huge caudex, 8: Ipomoea carnea, 9: Ipomoea batatas ‘Zhezishu 3’, 10: Ipomoea batatas ‘Eshu 6’, 11: Ipomoea batatas ‘Wanshu 7’ -
[1] SHEKHAR S, MISHRA D, BURAGOHAIN A K, et al. Comparative analysis of phytochemicals and nutrient availability in two contrasting cultivars of sweet potato (Ipomoea batatas L.)[J]. Food Chemistry, 2015, 173: 957-965. doi: 10.1016/j.foodchem.2014.09.172
[2] MOHANRAJ R, SIVASANKAR S. Sweet potato (Ipomoea batatas [L. ] Lam): A valuable medicinal food: A rievew[J]. Journal of Medicinal Food, 2014, 17(7): 733-741. doi: 10.1089/jmf.2013.2818
[3] YANG Z, ZHU P, KANG H, et al. High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.)[J]. BMC Genomics, 2020, 21(1). doi: 10.1186/s12864-020-6567-3.
[4] 谢一芝, 郭小丁, 贾赵东, 等. 中国食用甘薯育种现状及展望[J]. 江苏农业学报, 2018, 34(6): 1419-1424. doi: 10.3969/j.issn.1000-4440.2018.06.030 [5] 胡玲, 李强, 王欣, 等. 甘薯地方品种和育成品种的遗传多样性[J]. 江苏农业学报, 2010, 26(5): 925-935. doi: 10.3969/j.issn.1000-4440.2010.05.006 [6] 李强, 刘庆昌, 马代夫. 甘薯近缘野生种研究利用现状及展望[J]. 分子植物育种, 2006, 4(6S): 105-110. [7] 曹清河, 张安, 李鹏, 等. 甘薯近缘野生种的抗病性鉴定与新型种间杂种的获得[J]. 植物遗传资源学报, 2009, 10(2): 224-229. [8] GARRIDO-CARDENAS J A, MESA-VALLE C, MANZANO-AGUGLIARO F. Trends in plant research using molecular markers[J]. Planta, 2018, 247(3): 543-557. doi: 10.1007/s00425-017-2829-y
[9] YANG X S, SU W J, WANG L J, et al. Molecular diversity and genetic structure of 380 sweetpotato accessions as revealed by SSR markers[J]. Journal of Integrative Agriculture, 2015, 14(4): 633-641. doi: 10.1016/S2095-3119(14)60794-2
[10] 苏一钧, 王娇, 戴习彬, 等. 303份甘薯地方种SSR遗传多样性与群体结构分析[J]. 植物遗传资源学报, 2018, 19(2): 243-251. [11] 季志仙, 王美兴, 范宏环, 等. 基于ISSR指纹的甘薯食用品种的遗传多样性分析[J]. 核农学报, 2014, 28(7): 1197-1202. doi: 10.11869/j.issn.100-8551.2014.07.1197 [12] 王崇, 王连军, 苏文瑾, 等. 基于cpSSR标记的甘薯品种亲缘关系及遗传多样性分析[J]. 分子植物育种, 2020, 18(5): 1687-1696. [13] LEE K J, LEE G A, LEE J R, et al. Genetic diversity of sweet potato (Ipomoea batatas L. Lam) germplasms collected worldwide using chloroplast SSR markers[J]. Agronomy, 2019, 9(11): 725-740. doi: 10.3390/agronomy9110725
[14] GUALBERTO J M, NEWTON K J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation[M]//MERCHANT S S. Annual Review of Plant Biology: Vol 68. Palo alto: Annual Reviews. 2017: 225-252.
[15] PELLETIER G, BUDAR F. The molecular biology of cytoplasmically inherited male sterility and prospects for its engineering[J]. Current Opinion in Biotechnology, 2007, 18(2): 121-125. doi: 10.1016/j.copbio.2006.12.002
[16] LILLY J W, BARTOSZEWSKI G, MALEPSZY S, et al. A major deletion in the cucumber mitochondrial genome sorts with the MSC phenotype[J]. Current Genetics, 2001, 40(2): 144-151. doi: 10.1007/s002940100238
[17] QIN Y J, BUAHOM N, KROSCH M N, et al. Genetic diversity and population structure in Bactrocera correcta (Diptera: Tephritidae) inferred from mtDNA cox1 and microsatellite markers[J]. Scientific Reports, 2016, 6: 38476. doi: 10.1038/srep38476.
[18] BOUILLÉ M, SENNEVILLE S, BOUSQUET J. Discordant mtDNA and cpDNA phylogenies indicate geographic speciation and reticulation as driving factors for the diversification of the genus Picea[J]. Tree Genetics & Genomes, 2011, 7(3): 469-484.
[19] GODBOUT J, JARAMILLO-CORREA J P, BEAULIEU J, et al. A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range North American conifer[J]. Molecular Ecology, 2005, 14(11): 3497-3512. doi: 10.1111/j.1365-294X.2005.02674.x
[20] HU D, LUO Z. Polymorphisms of amplified mitochondrial DNA non-coding regions in Diospyros spp.[J]. Scientia Horticulturae, 2006, 109(3): 275-281. doi: 10.1016/j.scienta.2006.02.027
[21] 马丽, 周玉亮, 郝兆祥, 等. 石榴种质资源的matK基因序列分析[J]. 分子植物育种, 2020, 18(16): 5274-5279. [22] DUMINIL J, PEMONGE M H, PETIT R J. A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA[J]. Molecular Ecology Notes, 2002, 2(4): 428-430. doi: 10.1046/j.1471-8286.2002.00263.x
[23] DUMOLIN-LAPEGUE S, PEMONGE M H, PETIT R J. An enlarged set of consensus primers for the study of organelle DNA in plants[J]. Molecular Ecology, 1997, 6(4): 393-397. doi: 10.1046/j.1365-294X.1997.00193.x
[24] SUN X Q, ZHU Y J, GUO J L, et al. DNA barcoding the Dioscorea in China, a vital group in the evolution of monocotyledon: Use of matK gene for species discrimination[J]. PLoS One, 2012, 7(2): e32057. doi: 10.1371/journal.pone.0032057
[25] KUMAR S, STECHER G, LI M, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. doi: 10.1093/molbev/msy096
[26] ROZAS J, FERRER-MATA A, SÁNCHEZ-DELBARRIO J C, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets[J]. Molecular Biology and Evolution, 2017, 34(12): 3299-3302. doi: 10.1093/molbev/msx248
[27] 刘峥, 张汉尧. 旋花科植物ITS序列分析[J]. 西部林业科学, 2012, 41(4): 70-74. doi: 10.3969/j.issn.1672-8246.2012.04.012 [28] 俞立璇, 刘美艳, 曹清河, 等. 栽培种甘薯及其近缘野生种nrDNA ITS序列分析[J]. 植物科学学报, 2014, 32(1): 40-49. [29] JIN D P, LEE J H, XU B, et al. Phylogeography of East Asian Lespedeza buergeri (Fabaceae) based on chloroplast and nuclear ribosomal DNA sequence variations[J]. Journal of Plant Research, 2016, 129(5): 793-805. doi: 10.1007/s10265-016-0831-2
[30] 郭亚龙, 葛颂. 线粒体nad1基因内含子在稻族系统学研究中的价值: 兼论Porteresia的系统位置[J]. 植物分类学报, 2004, 42(4): 333-344. [31] 陆佳妮, 赵志礼, 倪梁红, 等. 线粒体nad1/b-c及nad5/d-e在秦艽组植物中物种鉴定意义的评价[J]. 药物学报, 2019, 54(1): 166-172. [32] SNOUSSI H, DUVAL M F, GARCIA-LOR A, et al. Assessment of the genetic diversity of the Tunisian citrus rootstock germplasm[J]. BMC Genetics, 2012: 13. doi: 10.1186/1471-2156-13-16.
[33] YANG J, VÁZQUEZ L, CHEN X D, et al. Development of chloroplast and nuclear DNA markers for Chinese Oaks (Quercus Subgenus Quercus) and assessment of their utility as DNA barcodes[J]. Frontiers in Plant Science, 2017: 8. doi: 10.3389/fpls.2017.00816.
[34] KORNELIUSSEN T S, MOLTKE I, ALBRECHTSEN A, et al. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data[J]. BMC Bioinformatics, 2013: 14. doi: 10.1186/1471-2105-14-289.



 下载:
下载: