Identification of autophagy-related genes in fall armyworm, Spodoptera frugiperda
-
摘要:目的
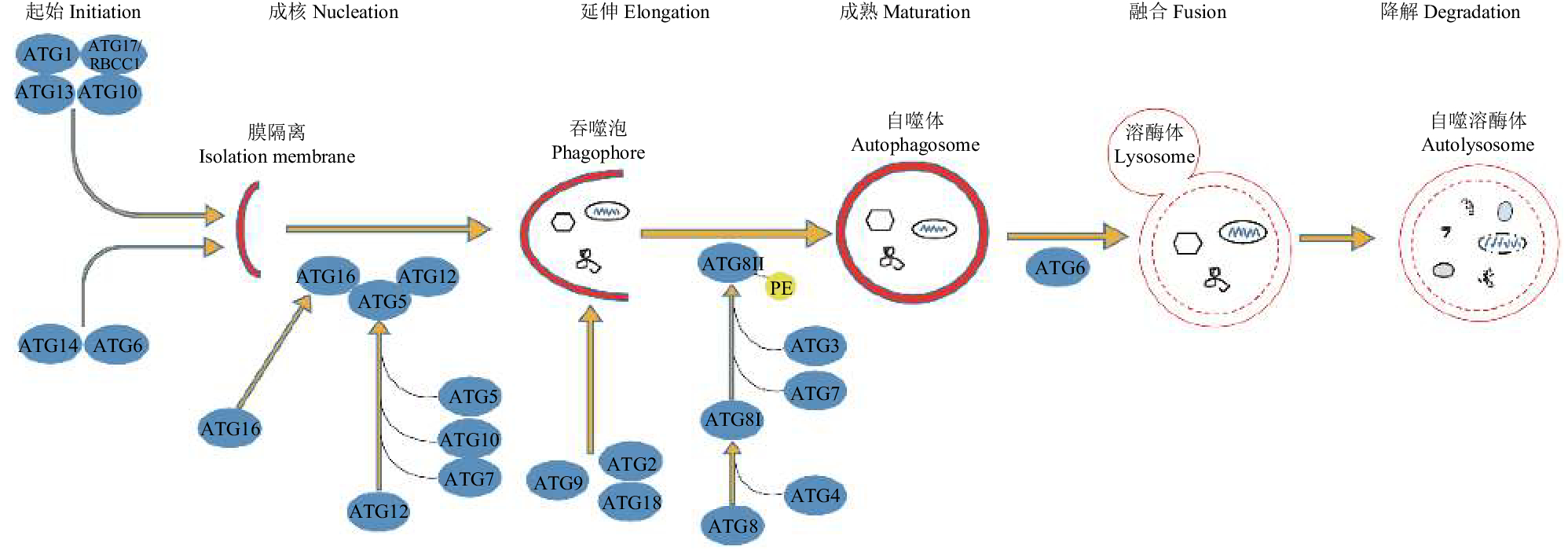
利用草地贪夜蛾Spodoptera frugiperda基因组序列,鉴定草地贪夜蛾自噬相关基因(Autophagy-related gene, Atg)及其对病毒感染的自噬应答,为研究自噬的分子机制奠定基础。
方法对人类Homo spiens、果蝇Drosophila melanogaster等不同物种来源的自噬相关蛋白(ATG)序列进行相似性比对,搜索草地贪夜蛾自噬相关基因序列,并通过NCBI保守结构域搜索工具预测候选自噬相关蛋白中存在的功能结构域;并通过qRT-PCR检测重组病毒EGFP-AcMNPV感染后引起Sf9细胞中自噬相关基因的表达。
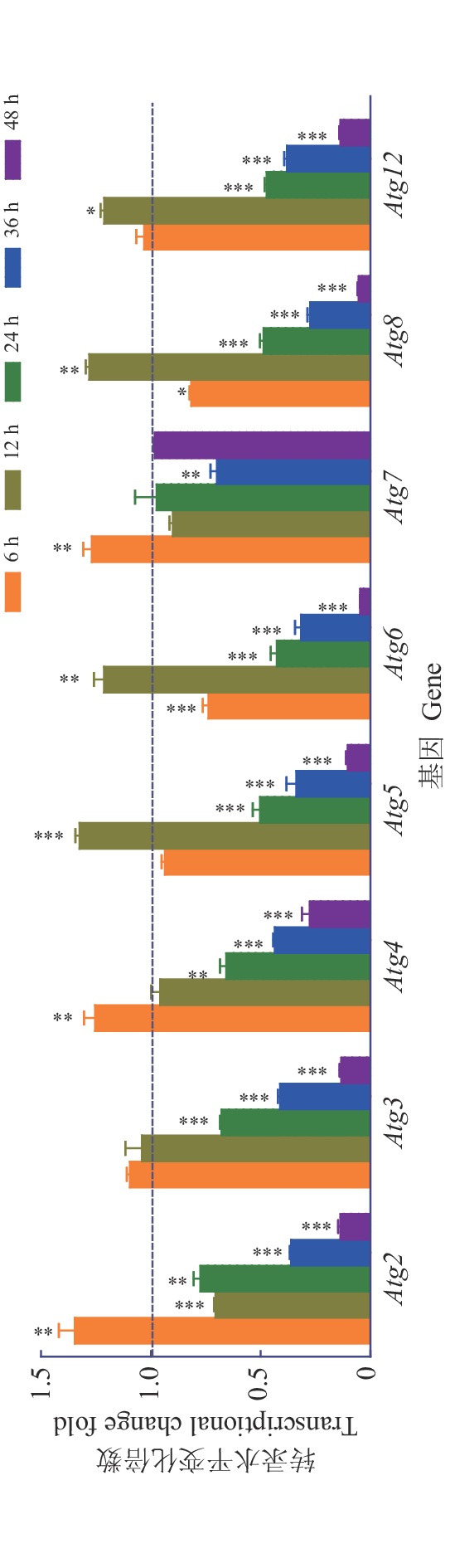
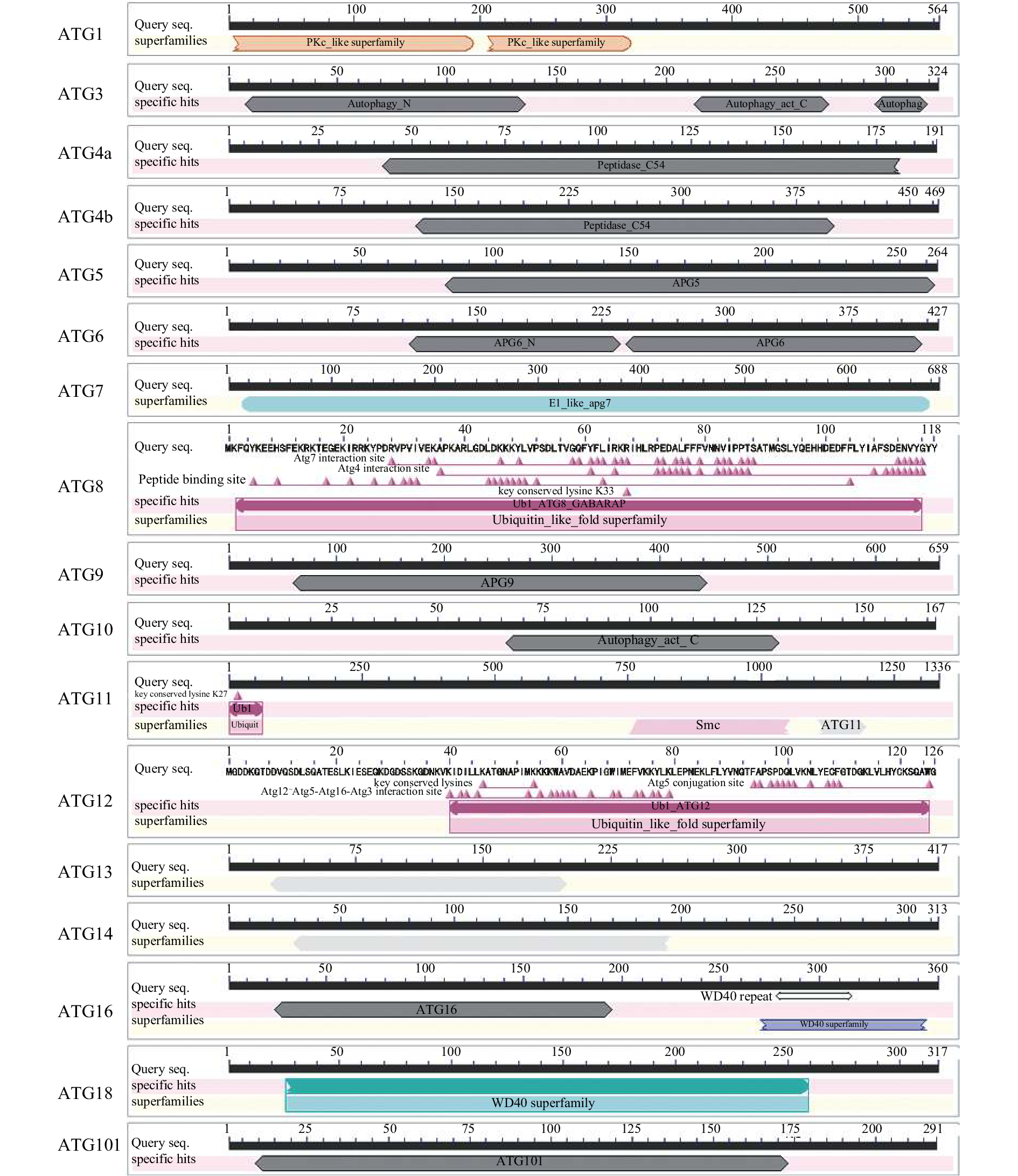
结果鉴定获得了草地贪夜蛾参与自噬体形成的核心Atg基因10多个,包括Atg3、Atg5、Atg6、Atg7、Atg10、Atg12、Atg13、Atg101的全序列以及Atg1、Atg2、Atg4、Atg9、Atg14、Atg16、Atg17、Atg18的部分序列,Atg2、Atg4、Atg5、Atg6、Atg7、Atg8、Atg12等自噬相关基因在重组病毒EGFP-AcMNPV感染Sf9细胞6~12 h后表达量上调。
结论获得草地贪夜蛾基因组中真核生物保守的自噬相关基因,暗示草地贪夜蛾的自噬途径在进化中是保守的。AcMNPV病毒感染Sf9细胞可能引发宿主细胞的自噬响应。
Abstract:ObjectiveTo identify autophagy-related genes (Atg) in fall armyworm (Spodoptera frugiperda) genome and their response to viral infection in S. frugiperda, and provide a basis for further research on the molecular mechanism of autophagy.
MethodAutophagy-related protein (ATG) sequences from various species, such as human (Homo sapiens) and fruit fly (Drosophila melanogaster), were aligned and analyzed for sequence similarity. ATG sequences in S. frugiperda genome were searched. Conservative domain (CD) of the candidate ATG were analyzed by NCBI CD search. Furthermore, qRT-PCR was used to detect the expression changes of Atgs in Sf9 cells after infection by recombinant EGFP-AcMNPV.
ResultWe identified more than ten core Atgs which were involved in autophagosome formation in S. frugiperda, including the complete sequences of Atg3, Atg5, Atg6, Atg7, Atg10, Atg12, Atg13 and Atg101, and the partial sequences of Atg1, Atg2, Atg4, Atg9, Atg14, Atg16, Atg17 and Atg18. The expression levels of S. frugiperda Atg2, Atg4, Atg5, Atg6, Atg7, Atg8 and Atg12 were up-regulated after 6 to 12 hours infection by EGFP-AcMNPV.
ConclusionEukaryotic conserved Atgs exist in the genome of S. frugiperda, indicating the autophagy pathway in S. frugiperda is evolutionarily conserved. AcMNPV infection might trigger the autophagic response in Sf9 cells.
-
-
图 5 AcMNPV对Sf9细胞对自噬相关基因转录水平的影响
图中数据为平均数±标准差,“*”、“**”、“***”分别表示相同基因在病毒感染后不同时间表达量在0.05、0.01和0.001水平差异显著(t检验)
Figure 5. Effects of AcMNPV on transcriptional levels of autophagy-related genes in Sf9 cells
Datum in the figure is mean ± standard deviation;“*”, “**” and “***” indicates significant differences at the levels of 0.05, 0.01 and 0.001 respectively (t test)
表 1 qRT-PCR引物序列
Table 1 Primer sequence for qRT-PCR
基因 Gene 正向引物(5′→3′) Forward primer 反向引物(5′→3′) Reverse primer Atg2 GATGTTTGCTCATGCCATAGACT ATGGACCTCAAGTGCGATGC Atg3 CGCGGAAGTAATGAAGAAAA CGTACTCGATGGTCGGGATG Atg4 TGTTGATAGTGCCGCTCAGG GTTGGGCTTACCACCGATCA Atg5 AACGACCCTCAACTACCGTG TGGACATGACTTGGCCTCTG Atg6 AAGGAGCAAGTTGAGAAAGGT ATTGAATAGGCATGAGGTGG Atg7 TTGGAAAGGAGCTGTGGACT CATACACGGTGGGCTGACTA Atg8 GAAGGCCAGGCTTGGAGACC GGCTGATGTTGGAGGAATGAC Atg12 ACAGGAAATGCCCCCATTATGA GGTCTGGAGATGGAGCAAATG ECD CTTCTGACCTGGCTACATTG ACCTGTTGTTTGTGGCTCTA 表 2 草地贪夜蛾自噬相关蛋白的序列与相似性分析
Table 2 Sequence and similarity analysis of Spodoptera frugiperda autophagy related proteins
自噬相关蛋白1) Autophagy-related protein 序列编号
Sequence no.结构域/蛋白家族2)
Domain/
Protein family黑腹果蝇 Drosophila melanogaster 人类 Homo sapiens 名称/NCBI登录号
Name/NCBI
accession number序列相似性/%
Sequence similarity名称/NCBI登录号
Name/NCBI
accession number序列相似性/%
Sequence similarityATG1* GSSPFG00023661001.3 PKc_like/
Protein kinasesAutophagy-related 1, isoform A/ NP_648601.1 69.79 Serine/threonine-protein kinase ULK2 / NP_001136082.1 64.10 ATG2* GSSPFG00005530001.3 Autophagy-related 2, isoform A/
NP_647748.130.96 Autophagy-related protein 2 homolog B/ NP_060506.6 32.29 ATG3 GSSPFG00021042001.3 Autophagy_N, _C and Autophay_act_C, Ubiquitin-like-conjugating enzyme ATG3/NP_649059.1 65.88 Ubiquitin-like-conjugating enzyme ATG3 isoform 1/ NP_071933.2 64.40 ATG4a* GSSPFG00032702001 Peptidase_C54/
Cysteine proteaseAutophagy-related 4a, isoform B/
NP_001259852.158.12 Cysteine protease ATG4B isoform b/ NP_847896.1 57.83 ATG4b* GSSPFG00022978001.3 Peptidase_C54/
Cysteine proteaseAutophagy-related 4b/
NP_650452.146.68 Cysteine protease ATG4D isoform 1 precursor/NP_116274.3 42.92 ATG5 GSSPFG00011494001.3 Autophagy protein Apg5 Autophagy-related 5/
NP_572390.158.49 Autophagy protein 5 isoform a/ NP_001273035.1 51.65 ATG6 GSSPFG00029332001.3 Autophagy protein Apg6 Autophagy-related 6/
NP_651209.156.53 Beclin-1 isoform a/NP_001300927.1 53.44 ATG7 GSSPFG00017981001.3 E1_like_Apg7 Autophagy-related 7, isoform A/
NP_611350.146.86 Ubiquitin-like modifier-activating enzyme ATG7 isoform X7/ XP_011531586.1 43.25 ATG8 GSSPFG00035793001.1 Ubiquitin-like _ATG8_GABARAP Autophagy-related 8a, isoform A/
NP_727447.193.10 Gamma-aminobutyric acid receptor-associated protein/ NP_009209.1 90.52 ATG9* GSSPFG00017231001.3
GSSPFG00013065001.3Autophagy protein Apg9 Autophagy-related 9, isoform A/ NP_611114.1 45.00 Autophagy-related protein 9A/NP_001070666.1 46.88 ATG10 GSSPFG00019539001.3 Autophagy_act_C Autophagy-related 10, isoform B/
NP_001097215.134.76 Ubiquitin-like-conjugating enzyme ATG10 isoform X2/ XP_005248669.1 33.53 ATG12 GSSPFG00008488001.3 Ubiquitin-like_
ATG12Autophagy-related 12/
NP_648551.354.55 Ubiquitin-like protein ATG12 isoform 1/NP_004698.3 69.57 ATG13 GSSPFG00023791001.3 Autophagy-related
protein 13Autophagy-related 13/
NP_649796.132.80 Autophagy-related protein 13 isoform g/NP_001333285.1 42.52 ATG14* GSSPFG00010270001.4 Atg14 super family Autophagy-related 14/
NP_651669.126.45 Beclin 1-associated autophagy-related key regulator/ NP_055739.2 24.12 ATG16* GSSPFG00003827001.3 Autophagy protein 16 and WD40 super family Autophagy-related 16, isoform C/
NP_733313.236.80 Autophagy-related protein 16-1 isoform 2/ NP_060444.3 31.37 ATG17* GSSPFG00021933001.4 Ubl_RB1CC1 and ATG11 domain-containing protein Autophagy-related 17, isoform B/
NP_996156.135.33 RB1-inducible coiled-coil protein 1 isoform X11/ XP_011515949.1 30.67 ATG18* GSSPFG00009401001.3 WD40 super family Autophagy-related 18a, isoform
F/NP_001261566.159.61 WD repeat domain phosphoinositide-interacting protein 2 isoform d/ NP_001028691.1 59.31 ATG101 GSSPFG00026235001.3 Autophagy-related protein 101 Autophagy-related 101/
NP_573326.156.11 Autophagy-related protein 101/ NP_068753.2 48.87 1)“*”表示获得该基因的部分序列; 2)早期在酵母中鉴定的参与自噬的基因被称为Apg,后来统一使用Atg来命名
1)“*”means partial sequence of this gene is obtained; 2)The early genes related to autophagy identified in yeast are called Apg, which are later uniformly named Atg -
[1] KUO C, HANSEN M, TROEMEL E. Autophagy and innate immunity: Insights from invertebrate model organisms[J]. Autophagy, 2018, 14(2): 233-242. doi: 10.1080/15548627.2017.1389824
[2] YIN Z, PASCUAL C, KLIONSKY D J. Autophagy: Machinery and regulation[J]. Microbial Cell, 2016, 3(12): 588-596. doi: 10.15698/mic2016.12.546
[3] SHENG R, QIN Z H. History and current status of autophagy research[J]. Advances in Experimental Medicine and Biology, 2019, 1206: 3-37.
[4] MATSUURA A, TSUKADA M, WADA Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae[J]. Gene, 1997, 192(2): 245-250. doi: 10.1016/S0378-1119(97)00084-X
[5] OHSUMI Y. Protein turnover[J]. International Union of Biochemistry and Moleular Biology: Life, 2006, 58(5/6): 363-369.
[6] SUZUKI H, OSAWA T, FUJIOKA Y, et al. Structural biology of the core autophagy machinery[J]. Current Opinion in Structural Biology, 2017, 43: 10-17. doi: 10.1016/j.sbi.2016.09.010
[7] PALMISANO N J, MELÉNDEZ A. Autophagy in C. elegans development[J]. Developmental Biology, 2019, 447(1): 103-125. doi: 10.1016/j.ydbio.2018.04.009
[8] YANO T, MITA S, OHMORI H, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila[J]. Nature Immunology, 2008, 9(8): 908-916. doi: 10.1038/ni.1634
[9] TALLOCZY Z, VIRGIN H T, LEVINE B. PKR-dependent autophagic degradation of herpes simplex virus type 1[J]. Autophagy, 2006, 2(1): 24-29. doi: 10.4161/auto.2176
[10] MARCHLER-BAUER A, BO Y, HAN L, et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures[J]. Nucleic Acids Research, 2017, 45(D1): D200-D203. doi: 10.1093/nar/gkw1129
[11] LUCKOW V A, LEE S C, BARRY G F, et al. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli[J]. Journal of Virology, 1993, 67(8): 4566-4579. doi: 10.1128/JVI.67.8.4566-4579.1993
[12] SHANG H, GARRETSON T A, KUMAR C M S, et al. Improved pFastBacTM donor plasmid vectors for higher protein production using the Bac-to-Bac® baculovirus expression vector system[J]. Journal of Biotechnology, 2017, 255: 37-46. doi: 10.1016/j.jbiotec.2017.06.397
[13] 余倩. 斜纹夜蛾核多角体病毒诱导细胞凋亡的研究[D]. 广州: 中山大学, 2008. [14] SALEM T Z, ALLAM W R, THIEM S M. Verifying the stability of selected genes for normalization in q-PCR experiments of Spodoptera frugiperda cells during AcMNPV infection[J]. PLoS One, 2014, 9(10): e108516. doi: 10.1371/journal.pone.0108516
[15] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method[J]. Methods, 2001, 25(4): 402-408. doi: 10.1006/meth.2001.1262
[16] 谢昆, 王丽仙, 王靖, 等. 草地贪夜蛾Sf9细胞对饥饿诱导自噬的响应[J]. 华北农学报, 2017, 32(3): 91-95. doi: 10.7668/hbnxb.2017.03.014 [17] 杨韵, 卢淼青, 张阳, 等. 高效氯氰菊酯对草地贪夜蛾Sf9细胞自噬的诱导作用[J]. 农药学学报, 2017, 19(2): 176-181. [18] VEERAN S, CUI G, SHU B, et al. Curcumin-induced autophagy and nucleophagy inSpodoptera frugiperda Sf9 insect cells occur via PI3K/AKT/TOR pathways[J]. Journal of Cell Biochemistry, 2018, 120(2): 2119-2137.
[19] CUI G, SHU B, VEERAN S, et al. Natural beta-carboline alkaloids regulate the PI3K/Akt/mTOR pathway and induce autophagy in insect Sf9 cells[J]. Pesticide Biochemistry and Physiology, 2019, 154: 67-77. doi: 10.1016/j.pestbp.2018.12.005
[20] REN C, FINKEL S E, TOWER J. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity[J]. Experimental Gerontology, 2009, 44(3): 228-235. doi: 10.1016/j.exger.2008.10.002
[21] VORONIN D, COOK D A, STEVEN A, et al. Autophagy regulates Wolbachia populations across diverse symbiotic associations[J]. Proceedings of the National Academy of Sciences, 2012, 109(25): E1638-E1646. doi: 10.1073/pnas.1203519109
[22] SHELLY S, LUKINOVA N, BAMBINA S, et al. Autophagy is an essential component of drosophila immunity against vesicular stomatitis virus[J]. Immunity, 2009, 30(4): 588-598. doi: 10.1016/j.immuni.2009.02.009
[23] NAKAMOTO M, MOY R H, XU J, et al. Virus recognition by toll-7 activates antiviral autophagy in Drosophila[J]. Immunity, 2012, 36(4): 658-667. doi: 10.1016/j.immuni.2012.03.003
[24] 张晓娟. 棉铃虫细胞自噬相关基因Atg8的分子特征以及对昆虫生长发育的影响[D]. 武汉: 华中师范大学, 2014. [25] 王霞. Atg6对棉铃虫繁殖的影响及其在昆虫细胞自噬中的作用[D]. 武汉: 华中师范大学, 2014. [26] YU F, HAO P, YE C, et al. NlATG1 gene participates in regulating autophagy and fission of mitochondria in the brown planthopper, Nilaparvata lugens[J]. Frontiers in Physiology, 2020, 10: 1622. doi: 10.3389/fphys.2019.01622



 下载:
下载: