Research progress of epigenetic mechanism of noncoding RNAs regulating avian skeletal muscle development
-
摘要:
肉、蛋、奶是畜牧业最主要的三大产品,其中,肉的需求量是最高的。肌肉是动物躯体的重要组成部分,仅骨骼肌就已占全身体质量的40%左右。骨骼肌在调节动物新陈代谢、机体运动、能量储存和健康等方面至关重要,是机体功能正常运转的必要组分。骨骼肌发育过程极其复杂,主要包括体节细胞增殖分化、成肌细胞增殖分化、肌管成熟以及肌纤维形成等环节,整个过程受许多遗传因子调控,其中,由微小RNAs(miRNAs)、长链非编码RNAs(lncRNAs)、环状RNAs(circRNAs)等几种类型构成的非编码RNAs(ncRNAs)可以通过靶向关键因子调控骨骼肌发育过程。本文介绍了各类ncRNAs的特征与功能,总结了近年来有关ncRNAs在家禽肌肉生长发育中的研究,阐述ncRNAs在骨骼肌生长发育进程中的表观遗传调控机制,为改善家禽生长发育提供参考。
Abstract:Meat, eggs and milk are three important products in animal husbandry, among which, the demand for meat is the highest. Muscle is an essential component of animal body, and skeletal muscle accounts for about 40% of body weight. Skeletal muscle plays an important role in animal metabolism, body movement, energy storage and health, and it's the essential part in body function normal running. The development process of skeletal muscle is extremely complex, mainly including somite proliferation and differentiation, myoblast proliferation and differentiation, myotube fusion, and the formation of muscle fiber, and the whole progress is regulated by many genetic factors. Non-coding RNAs (ncRNAs), which mainly consist of micro RNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), can regulate skeletal muscle development by targeting key factors. This paper briefly described the characteristics and functions of those ncRNAs, then reviewed recent studies of ncRNAs in avian muscle growth and development, and elucidated the epigenetic regulatory mechanism of ncRNAs in skeletal muscle growth and development, which could provide references for improving avian growth and development.
-
Keywords:
- noncoding RNA /
- skeletal muscle /
- growth and development /
- epigenetic regulation
-
小麦是仅次于大米的主要粮食作物,是人类生活中不可缺少的食物,通常利用制粉设备加工成面粉,制作成面包、面条等各种面食供人类食用。全世界每年需要加工制粉的谷物有20多亿t,其中小麦大约有6亿t[1]。辊式磨粉机是加工面粉的主要设备,其工作原理是通过1对水平排列并以不同角速度高速相向旋转的圆柱形磨辊,对小麦粉料施加挤压、剪切、摩擦等方式的载荷,将物料颗粒压碎、研磨成细粉,制成各种不同用途的成品面粉[2-3]。
磨辊是辊式磨粉机的核心部件,其内层材料以灰口铁为主,外层抗磨部分主要为白口铁。制粉过程中其与小麦粉料剧烈摩擦,产生磨料磨损现象,造成磨辊表面原有的形态发生变化,如齿辊的齿部钝化、光辊表面粗糙度降低。当磨辊磨损达到一定程度后,磨粉机出粉率与生产效率都会明显降低,需重新做磨光拉丝或喷砂处理[4-6]。磨辊磨损问题已经成为制约辊式磨粉机发展和应用的瓶颈,提高磨辊表面耐磨性能是目前制粉行业急需解决的难题。因此,磨辊金属材料的耐磨性是非常重要的性能,金属材料的耐磨性与表面硬度之间存在相应的关系,一般情况下,同类材料硬度增大则耐磨性提高,因此可以通过表面硬度间接反映材料的耐磨性[7-8]。
激光表面淬火是强化材料表面硬度的一种热处理方法[9-10],该技术解决了许多普通热处理工艺无法解决的难题,广泛用于汽车、冶金、模具、五金、轻工、机械制造等行业[11-17]。有学者研究了激光淬火工艺参数对HT210、模具钢718等材料淬硬层深度及表面硬度的影响[18-21],发现获得高而均匀的硬度是提高铸铁材料耐磨性的关键。本研究拟利用响应曲面方法设计激光淬火试验,探究激光淬火工艺参数对磨辊表面金属材料硬度的影响规律,并确定最优工艺参数组合,探讨经激光淬火处理后磨辊表面金属材料的性能变化和磨损机理。
1. 材料与方法
1.1 材料
试验材料为低铬白口铁,尺寸为57 mm × 25.5 mm × 6 mm,表面机械研磨抛光,其化学成分质量分数如下:C 2.6%~3.2%、Si 低于0.8%、Mn 1.0%~2.5%、Cr 2.0%~3.0%、Mo 2.0%~3.0%、Cu 2.0%~3.0%。
磨损试验中使用的磨料为甘肃产‘西旱1号’小麦籽粒,自然风干后经破碎、筛分及匀化处理制备粒度分布为0.5~1.5 mm的小麦粉料。小麦粉料不同成分质量分数为:淀粉71.8%、粗蛋白12.9%、水分9.8%、脂肪2.2%、粗纤维1.7%、粗灰分1.6%。
1.2 试验设备
1.2.1 激光淬火试验
使用额定功率200 W,波长1 070 nm的光纤激光器,工作频率40 Hz,脉宽20 ms。
1.2.2 硬度测定试验与金相组织试验
硬度测定采用莱州华银公司生产的HVS−1000型数显显微硬度计,施加载荷100 g,加载时间10 s。每组试样硬度测定试验重复3次,取3次测量值的平均值。采用MR5000型倒置金相显微镜(南京江南永新光学公司生产)检验试样金相组织。
1.2.3 磨损试验
采用MLS−225型橡胶轮式磨损试验机(张家口市宣化科华试验机制造有限公司生产)进行三体磨料磨损试验,采用精度为0.1 mg的分析天平称量试样磨损前后的质量损失,采用扫描电子显微镜(东莞市天测光学设备有限公司生产)观察试样被磨面的表面微观形貌。
1.3 试验方法
1.3.1 激光淬火试验
试样编号为1~20,分别进行不同工艺参数的激光表面淬火热处理,表面淬火区域的淬火扫描点呈线性排列在试样表面,如图1,淬火面积为30 mm × 18 mm。在进行激光淬火试验前,将试样置入丙酮溶液中,放入清洗机清洗6 min,用碳素吸光涂料对试样作黑化处理,提高材料对激光的吸收率。完成激光淬火试验后,将试样沿着与激光扫描垂直的方向切开,用硬度计测量硬度值。
1.3.2 响应曲面试验设计
通过前期单因素激光淬火试验筛选,影响磨辊表面硬度的因素主要是激光功率、光斑直径和扫描速度。根据中心复合的旋转组合设计原理,以激光功率(A)、光斑直径(B)、扫描速度(C)为试验影响因素,以硬度(R)作为响应指标,采用3因素5水平试验。各试验因素水平如表1所示。
表 1 因素水平表Table 1. Factor-level table水平
Level激光功率/W
Laser power光斑直径/mm
Spot diameter扫描速度/(mm·s−1)
Scanning speed−1.682 163 0.53 166 −1 170 0.60 200 0 180 0.70 250 +1 190 0.80 300 +1.682 197 0.87 334 1.3.3 磨损试验
在三体磨料磨损试验前,磨损试验机的参数设定如下:转速400 r/min,压力225 N,轧距0.15 mm。选用硬度为60 邵尔的橡胶轮,在室温条件下分别对未经激光淬火处理、经激光淬火最优工艺参数组合处理的2组试样进行为期2 h的抗小麦粉料磨损试验,共计5个磨损周期,即总磨程为10 h。将试样磨损前后的质量损失作为评价指标,每组试样磨损试验重复3次,取3次测量值的平均值作为分析数据。
1.4 数据统计与分析
采用Design-expert 8.0.6对数据进行统计分析;用Origin 8.0软件进行作图。
2. 结果与分析
2.1 激光淬火试验
响应面各因素试验设计与结果如表2所示。使用Design-expert 8.0.6软件分析试验结果,得到各因素与响应值R的二次回归方程:
表 2 试验设计与结果Table 2. Experiment design and result试样编号
Sample number因素 Factor 硬度/HV
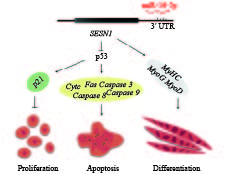
Hardness激光功率 Laser power 光斑直径 Spot diameter 扫描速度 Scanning speed 1 −1 −1 −1 545.36 2 1 −1 −1 600.13 3 −1 1 −1 520.86 4 1 1 −1 637.81 5 −1 −1 1 530.77 6 1 −1 1 515.00 7 −1 1 1 592.02 8 1 1 1 560.35 9 −1.682 0 0 532.10 10 +1.682 0 0 695.13 11 0 −1.682 0 534.76 12 0 +1.682 0 658.30 13 0 0 −1.682 627.76 14 0 0 +1.682 521.55 15 0 0 0 644.24 16 0 0 0 680.32 17 0 0 0 659.18 18 0 0 0 663.26 19 0 0 0 687.08 20 0 0 0 631.29 $$\begin{aligned} {{R}} = & {{661}}{{.80}} + {{2}}9.18{{A}} + {{2}}3.98{{B}} - {{2}}0.84{{C}} + \\ & {{5}}{{.79AB}} - {{2}}7.39{{AC}} + {{1}}1.68{{BC}} - \\ & {{22}}{{.66}}{{{A}}^{{2}}} - {{2}}8.70{{{B}}^{{2}}} - {{3}}6.43{{{C}}^{{2}}} {\text{。}} \end{aligned}$$ 对回归方程进行方差分析和回归系数显著性检验,结果见表3。回归模型P<0.01,表明回归模型中各因素与响应值的相关性是显著的。其中模型的一次项A(激光功率)、B(光斑直径)与C(扫描速度)对磨辊材料表面硬度影响显著(P<0.05);二次项B2、C2对磨辊材料表面硬度影响极显著(P<0.01),A2影响显著(P<0.05);交互项AC对磨辊材料表面硬度影响显著(P<0.05),AB与BC影响均不显著(P>0.05)。根据模型各因素回归系数和P值大小,得到影响磨辊材料表面硬度的各因素依次为激光功率、光斑直径、扫描速度。模型的复相关系数为0.846 8,模型的校正决定系数Radj2为0.708 9,试验误差小,可以用于硬度的预测。
表 3 方差分析表1)Table 3. Variance analysis table变异来源 Variance source SS DF MS F P 模型 Model 65 184.71 9 7 242.75 6.14 0.004 5 A 11 625.85 1 11 625.85 9.86 0.010 5 B 7 855.99 1 7 855.99 6.66 0.027 4 C 5 932.68 1 5 932.68 5.03 0.048 8 AB 267.73 1 267.73 0.23 0.644 0 AC 6 003.89 1 6 003.89 5.09 0.047 7 BC 1 090.91 1 1 090.91 0.93 0.358 8 A2 7 396.70 1 7 396.70 6.27 0.031 2 B2 11 866.84 1 11 866.84 10.06 0.010 0 C2 19 125.48 1 19 125.48 16.22 0.002 4 残差 Residual 11 792.58 10 1 179.26 失拟项 Lack of fit 9 567.22 5 1 913.44 4.30 0.067 7 纯误差 Pure error 2 225.36 5 445.07 总计 Total 76 977.29 19 1)A、B、C分别为激光功率、光斑直径、扫描速度
1) A, B and C indicated laser power, spot diameter and scanning speed, respectively2.2 淬火工艺参数交互作用分析
为了考察各因素及其交互作用对磨辊材料表面硬度的影响,采用Design-expert 8.0.6软件得到了各因素间的响应曲面图和等高线图,如图2~4所示。各因素间交互作用的显著性取决于响应曲面的陡峭程度。响应曲面坡度越陡,说明该因素对响应值的影响越显著。当等高线的形状为椭圆形时,线密度大,表明因素间交互作用对硬度影响显著;等高线的形状为圆形或近似圆形时,线密度小,交互作用对硬度影响不显著。
2.2.1 激光功率与扫描速度对硬度的影响
激光功率方向的坡度比扫描速度陡峭(图2a),表明激光功率对磨辊材料表面硬度的影响大于扫描速度。等高线图形状呈椭圆形(图2b),表明激光功率与扫描速度间的交互作用对硬度影响显著。
2.2.2 激光功率与光斑直径对硬度的影响
响应面图中激光功率方向的曲线坡度大于光斑直径方向(图3a),说明激光功率对磨辊材料表面硬度的影响大于光斑直径。与激光功率和扫描速度交互作用下等高线密度(图2b)相比较,图3b中的等高线轮廓近似圆形,线密度较小,表明激光功率与光斑直径间的交互作用对硬度影响不显著。
2.2.3 光斑直径与扫描速度对硬度的影响
从响应曲面图可观察出,光斑直径方向响应面曲线比扫描速度方向陡峭(图4a),表明光斑直径对磨辊材料表面硬度的影响大于扫描速度。等高线图的线密度小,轮廓呈圆形(图4b),说明光斑直径与扫描速度间的交互作用对硬度的影响也不显著,这与回归分析结果一致。由上述结果可知,影响磨辊材料表面硬度的最主要因素为激光功率,其次为光斑直径和扫描速度。
2.2.4 激光淬火工艺参数最优组合
在实际面粉生产中,最终目的是提高磨辊的硬度,加强磨辊的耐磨性能,延长磨辊的使用周期。本试验利用响应曲面旋转二次组合设计方法,采用激光功率、光斑直径和扫描速度3个参数的试验范围作为约束条件,经过显著性检验的响应值R作为目标函数,经过非线性优化后得出最优的参数组合。分析得到激光淬火优化参数组合为:激光功率190 W,光斑直径0.74 mm,扫描速度220.14 mm/s,该参数组合下的试样表面硬度为688.67 HV。考虑到实际试验操作的便利,将此工艺条件进行进一步修正,得到可在实际生产中应用的工艺参数组合:激光功率190 W,光斑直径0.70 mm,扫描速度220 mm/s。为了检验软件分析结果的正确性,用上述最佳的淬火工艺参数进行3次验证试验,试验的结果与软件分析的结果基本吻合。
2.3 金相组织分析
该试样的原始硬度为509 HV,激光淬火处理后其硬度提升了35%,对试样进行激光淬火处理提升其耐磨性的本质是使其金相组织发生变化。图5为试样淬火后的金相组织图。
在激光快速加热条件下奥氏体晶粒非常细小。快速加热升温增加了奥氏体内碳、铬等元素的溶解度。碳化物在奥氏体内溶解,使其薄弱处发生断裂,形态得到改善。碳、铬等元素在奥氏体内溶解使得它们在奥氏体内的溶入量增长,激光淬火后得到的马氏体含碳量增加,基体硬度提高。同时,激光淬火使试样内部组织晶粒细化,形成大量马氏体,残留少量奥氏体。因此,试样经激光淬火后耐磨性得到极大的改善。试样在淬火后未产生裂纹现象。
2.4 磨损试验
2.4.1 质量损失试验结果
小麦粉料与试样表面接触时,其中的硬颗粒会与试样表面发生摩擦,试样表面因塑性挤压产生划痕,同时试样表面因压入的粉料硬颗粒形成沟槽,试样表面经多次塑性变形,发生疲劳破坏,表面材料掉落,脱离母体,造成试样质量损失。图6所示为未经激光淬火处理和激光淬火最优参数组合处理2组试样以小麦粉料为磨料的三体磨料磨损试验质量损失。对比图6中2组数据可知,经激光淬火最优参数组合处理后的试样质量损失约为未经激光淬火处理的试样的7%,由此可知,激光淬火处理后磨辊表面材料抗小麦粉料磨损性能显著提升。
2.4.2 磨损面微观形貌分析
图7所示为激光淬火前后2组试样典型被磨表面微观形貌扫描电子显微镜图。试样经激光淬火处理后,小麦粉料在试样表面的划痕较轻,粉料中的坚硬颗粒在激光淬火试样表面很难存留,表面上形成的划痕比较短,在小麦粉料作用下形成的沟槽更浅窄。因此,经激光淬火后的磨辊表面材料与小麦粉料间的摩擦磨损作用减弱,塑性变形次数降低,磨辊表面疲劳破坏得到改善,磨辊的磨损周期延长,生产成本降低。
3. 结论
本文采用旋转组合设计方法设计3因素5水平响应曲面试验,探究了激光功率、光斑直径和扫描速度对磨辊金属材料硬度的影响规律,并对3个试验因素进行参数优化,探讨磨辊材料耐磨性变化,得到以下结论:
1)各激光淬火工艺参数对磨辊金属材料硬度影响依次为激光功率>光斑直径>扫描速度;激光功率与扫描速度间的交互作用对硬度影响显著。
2)提高磨辊表面材料硬度的最优激光淬火工艺参数组合为:激光功率190 W、光斑直径0.70 mm、扫描速度220 mm/s;激光淬火处理后试样硬度提升了35%。
3)经激光淬火最佳工艺参数组合处理后试样的质量损失约为未经激光淬火处理试样的7%,经激光淬火处理后的磨辊表面材料与小麦粉料间的摩擦磨损作用减弱,小麦粉料在试样表面的划痕较轻较短,试样磨损面的沟槽更浅窄。
本试验结果表明,经过激光淬火处理后磨辊表面材料较未经处理的材料硬度显著提升,耐磨性能增强,这与华希俊等[22]的研究结果一致。淬火后的试样与小麦粉料发生摩擦磨损时,由于其表面的硬度得到强化,小麦粉料中的硬颗粒很难存留在试样表面,在表面产生的划痕与沟槽极为浅窄,使材料表面的损伤减少,极大地缓解了磨辊磨损严重等问题。
-
-
[1] 杨宁. 家禽业的核心将是保持高效优势[J]. 北方牧业, 2018(15): 7. [2] GÜLLER I, RUSSELL A P. MicroRNAs in skeletal muscle: Their role and regulation in development, disease and function[J]. J Physiol, 2010, 588(21): 4075-4087.
[3] PEARSON A M. Muscle growth and exercise[J]. Crit Rev Food Sci Nutr, 1990, 29(3): 167-196. doi: 10.1080/10408399009527522
[4] FENG Y, CAO J H, LI X Y, et al. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts[J]. Cell Biochem Funct, 2011, 29(5): 378-383. doi: 10.1002/cbf.1760
[5] SCHIAFFINO S, SANDRI M, MURGIA M. Activity-dependent signaling pathways controlling muscle diversity and plasticity[J]. Physiology (Bethesda), 2007, 22: 269-278.
[6] ZANOU N, GAILLY P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways[J]. Cell Mol Life Sci, 2013, 70(21): 4117-4130. doi: 10.1007/s00018-013-1330-4
[7] RULLMAN E, FERNANDEZ-GONZALO R, MEKJAVIĆ I B, et al. MEF2 as upstream regulator of the transcriptome signature in human skeletal muscle during unloading[J]. Am J Physiol Regul Integr Comp Physiol, 2018, 315(4): R799-R809. doi: 10.1152/ajpregu.00452.2017
[8] BRAUN T, GAUTEL M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis[J]. Nat Rev Mol Cell Biol, 2011, 12(6): 349-361. doi: 10.1038/nrm3118
[9] PERRY R L S, RUDNICK M A. Molecular mechanisms regulating myogenic determination and differentiation[J]. Front Biosci, 2000, 5: D750-D767. doi: 10.2741/A548
[10] GAO P F, GUO X H, DU M, et al. LncRNA profiling of skeletal muscles in Large White pigs and Mashen pigs during development[J]. J Anim Sci, 2017, 95(10): 4239-4250. doi: 10.2527/jas2016.1297
[11] CAO Y, YOU S, YAO Y, et al. Expression profiles of circular RNAs in sheep skeletal muscle[J]. Asian-Australas J Anim Sci, 2018, 31(10): 1550-1557. doi: 10.5713/ajas.17.0563
[12] SHI L, ZHOU B, LI P, et al. MicroRNA-128 targets myostatin at coding domain sequence to regulate myoblasts in skeletal muscle development[J]. Cell Signal, 2015, 27(9): 1895-1904. doi: 10.1016/j.cellsig.2015.05.001
[13] DINGER M E, PANG K C, MERCER T R, et al. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities[J]. PloS Comput Biol, 2008, 4(11). doi: 10.1371/journal.pcbi.1000176.
[14] CESANA M, CACCHIARELLI D, LEGNINI I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA[J]. Cell, 2011, 147(2): 358-369. doi: 10.1016/j.cell.2011.09.028
[15] OUYANG H, CHEN X, WANG Z, et al. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens[J]. DNA Res, 2018, 25(1): 71-86. doi: 10.1093/dnares/dsx039
[16] BALLARINO M, MORLANDO M, FATICA A, et al. Non-coding RNAs in muscle differentiation and musculoskeletal disease[J]. J Clin Invest, 2016, 126(6): 2021-2030. doi: 10.1172/JCI84419
[17] HAYES J, PERUZZI P P, LAWLER S. MicroRNAs in cancer: Biomarkers, functions and therapy[J]. Trends Mol Med, 2014, 20(8): 460-469. doi: 10.1016/j.molmed.2014.06.005
[18] LUO W, NIE Q, ZHANG X. MicroRNAs involved in skeletal muscle differentiation[J]. J Genet Genomics, 2013, 40(3): 107-116. doi: 10.1016/j.jgg.2013.02.002
[19] SAUNDERS M A, LIANG H, LI W H. Human polymorphism at microRNAs and microRNA target sites[J]. Proc Natl Acad Sci USA, 2007, 104(9): 3300-3305. doi: 10.1073/pnas.0611347104
[20] LAI E C. Micro RNAs are complementary to 3′UTR sequence motifs that mediate negative post-transcriptional regulation[J]. Nat Genet, 2002, 30(4): 363-364. doi: 10.1038/ng865
[21] SEOK H, HAM J, JANG E S, et al. MicroRNA target recognition: Insights from transcriptome-wide non-canonical interactions[J]. Mol Cells, 2016, 39(5): 375-381. doi: 10.14348/molcells.2016.0013
[22] LIU G, ZHANG R, XU J, et al. Functional conservation of both CDS- and 3′-UTR-located MicroRNA binding sites between species[J]. Mol Biol Evol, 2015, 32(3): 623-628.
[23] KREK A, GRÜN D, POY M N, et al. Combinatorial microRNA target predictions[J]. Nat Genet, 2005, 37(5): 495-500. doi: 10.1038/ng1536
[24] GIRAL H, KRATZER A, LANDMESSER U. MicroRNAs in lipid metabolism and atherosclerosis[J]. Best Pract Res Clin Endocrinol Metab, 2016, 30(5): 665-676. doi: 10.1016/j.beem.2016.11.010
[25] GROSS N, KROPP J, KHATIB H. MicroRNA signaling in embryo development[J]. Biology, 2017, 6(3). doi: 10.3390/biology6030034.
[26] 贾新正. 快慢型肉鸡miRNA的表达谱分析[D]. 广州: 华南农业大学, 2010. [27] WANG X G, YU J F, ZHANG Y, et al. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle[J]. Poult Sci, 2012, 91(1): 139-149. doi: 10.3382/ps.2011-01656
[28] LIN S, LI H, MU H, et al. Let-7b regulates the expression of the growth hormone receptor gene in deletion-type dwarf chickens[J]. BMC Genomics, 2012, 13. doi: 10.1186/1471-2164-13-306.
[29] WANG X G, SHAO F, GONG D Q, et al. miR-133a targets BIRC5 to regulate its gene expression in chicken[J]. Scientia Agricultura Sinica, 2013, 46(7): 1441-1447.
[30] OUYANG H, HE X, LI G, et al. Deep sequencing analysis of miRNA expression in breast muscle of fast-growing and slow-growing broilers[J]. Int J Mol Sci, 2015, 16(7): 16242-16262. doi: 10.3390/ijms160716242
[31] LUO W, WU H, YE Y, et al. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation[J]. Cell Death Dis, 2014, 5. doi: 10.1038/cddis.2014.289.
[32] WANG Z, OUYANG H, CHEN X, et al. Gga-miR-205a affecting myoblast proliferation and differentiation by targeting CDH11[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00414.
[33] TOWNLEY-TILSON W H D, CALLIS T E, WANG D Z. MicroRNAs 1, 133, and 206: Critical factors of skeletal and cardiac muscle development, function, and disease[J]. Int J Biochem Cell Biol, 2010, 42(8): 1252-1255. doi: 10.1016/j.biocel.2009.03.002
[34] JIA X, LIN H, ABDALLA B A, et al. Characterization of miR-206 promoter and its association with birthweight in chicken[J]. Int J Mol Sci, 2016, 17(4). doi: 10.3390/ijms17040559
[35] LI G, LUO W, ABDALLA B A, et al. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1[J]. Cell Death Dis, 2017, 8. doi: 10.1038/cddis.2017.479.
[36] GUO L, HUANG W, CHEN B, et al. Gga-mir-133a-3p regulates myoblasts proliferation and differentiation by targeting PRRX1[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00577.
[37] WANG Z, ZHANG X, LI Z, et al. MiR-34b-5p mediates the proliferation and differentiation of myoblasts by targeting IGFBP2[J]. Cells, 2019, 8(4). doi: org/10.3390/cells8040360.
[38] WANG J, HELIN K, JIN P, et al. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1[J]. Cell Growth Differ, 1995, 6(10): 1299-1306.
[39] LUO W, LI G, YI Z, et al. E2F1-miR-20a-5p/20b-5p auto-regulatory feedback loop involved in myoblast proliferation and differentiation[J]. Sci Rep. 2016, 6. doi: 10.1038/srep27904.
[40] JIA X, OUYANG H, ABDALLA B A, et al. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities[J]. Biochim Biophys Acta Gene Regul Mech, 2017, 1860(6): 674-684. doi: 10.1016/j.bbagrm.2017.02.010
[41] JIA X, LIN H, NIE Q, et al. A short insertion mutation disrupts genesis of miR-16 and causes increased body weight in domesticated chicken[J]. Sci Rep, 2016, 6. doi: 10.1038/srep36433.
[42] YANG Y L, LOH K S, LIOU B Y, et al. SESN-1 is a positive regulator of lifespan in Caenorhabditis elegans[J]. Exp Gerontol, 2013, 48(3): 371-379. doi: 10.1016/j.exger.2012.12.011
[43] EL HUSSEINI N, HALES B F. The roles of P53 and its family proteins, P63 and P73, in the DNA damage stress response in organogenesis stage mouse embryos[J]. Toxicol Sci, 2018, 162(2): 439-449. doi: 10.1093/toxsci/kfx270
[44] CAI B, MA M, CHEN B, et al. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation[J]. Cell Death Dis, 2018, 9. doi: 10.1038/s41419-018-0403-6.
[45] CABILI M N, TRAPNELL C, GOFF L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses[J]. Genes Dev, 2011, 25(18): 1915-1927.
[46] OKAZAKI Y, FURUNO M, KASUKAWA T, et al. Analysis of the mouse transcriptome based on functional annotation of 60, 770 full-length cDNAs[J]. Nature, 2002, 420(6915): 563-573. doi: 10.1038/nature01266
[47] WILUSZ J E, SUNWOO H, SPECTOR D L. Long noncoding RNAs: Functional surprises from the RNA world[J]. Genes Dev, 2009, 23(13): 1494-1504. doi: 10.1101/gad.1800909
[48] SANLI I, LALEVÉE S, CAMMISA M, et al. Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis[J]. Cell Rep, 2018, 23(2): 337-348. doi: 10.1016/j.celrep.2018.03.044
[49] KALLEN A N, ZHOU X B, XU J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs[J]. Mol Cell, 2013, 52(1): 101-112. doi: 10.1016/j.molcel.2013.08.027
[50] ZHOU L, SUN K, ZHAO Y, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1[J]. Nat Commun, 2015, 6. doi: 10.1038/ncomms10026.
[51] LI T, WANG S, WU R, et al. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing[J]. Genomics, 2012, 99(5): 292-298. doi: 10.1016/j.ygeno.2012.02.003
[52] LI Z, OUYANG H, ZHENG M, et al. Integrated analysis of long non-coding RNAs (lncRNAs) and mRNA expression profiles reveals the potential role of lncRNAs in skeletal muscle development of the chicken[J]. Front Physiol, 2017, 7. doi: 10.3389/fphys.2016.00687.
[53] OUYANG H, WANG Z, CHEN X, et al. Proteomic analysis of chicken skeletal muscle during embryonic development[J]. Front Physiol, 2017, 8. doi: 10.3389/fphys.2017.00281.
[54] LI Z, CAI B, ABDALLA B A, et al. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway[J]. J Cachexia Sarcopenia Muscle, 2019, 10(2): 391-410. doi: 10.1002/jcsm.v10.2
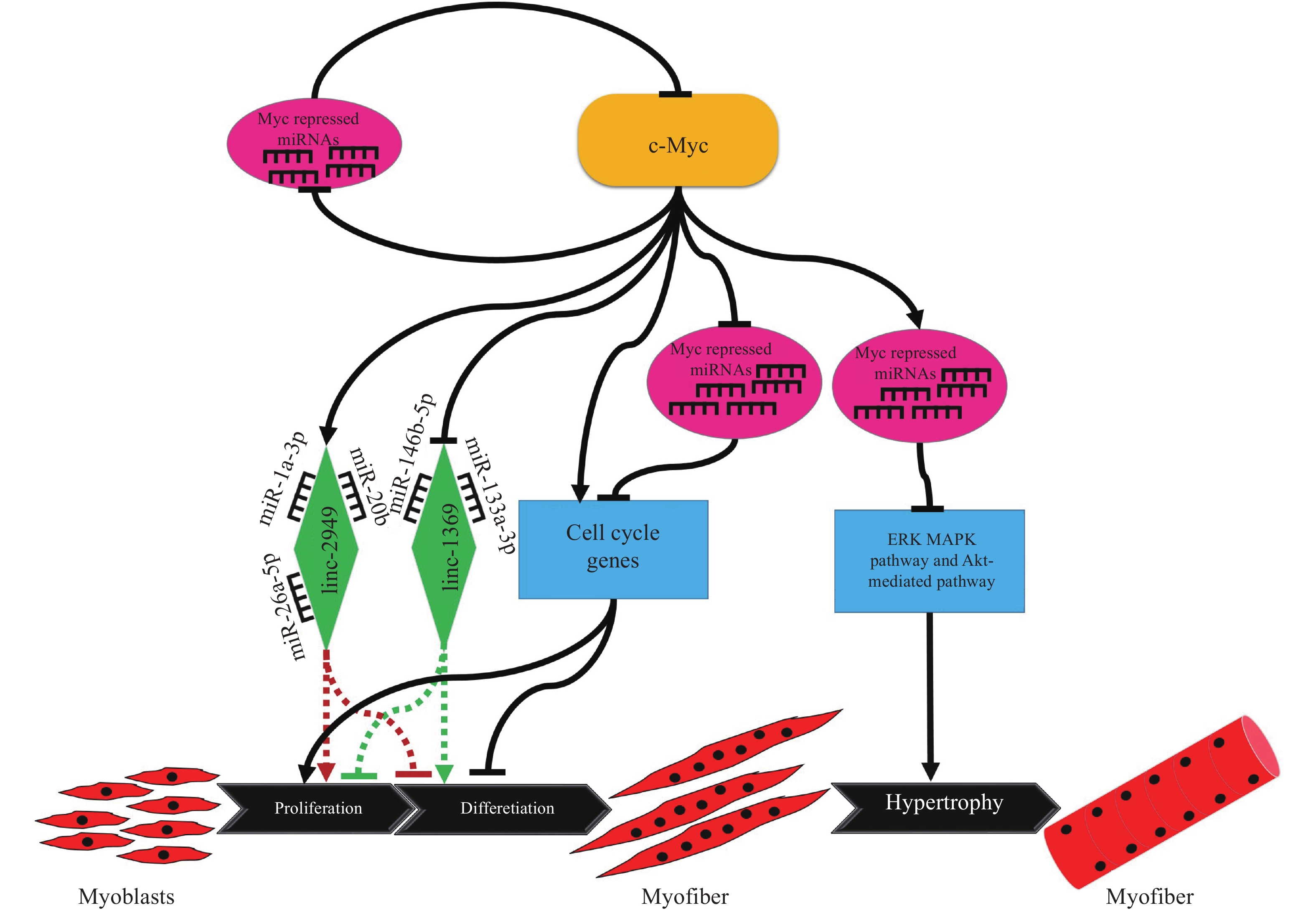
[55] LUO W, CHEN J, LI L, et al. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs[J]. Cell Death Differ, 2019, 26(3): 426-442. doi: 10.1038/s41418-018-0129-0
[56] CAI B, LI Z, MA M, et al. LncRNA-Six1 encodes a micropeptide to activate Six1 in cis and is involved in cell proliferation and muscle growth[J]. Front Physiol, 2017, 8. doi: 10.3389/fphys.2017.00230.
[57] MA M, CAI B, JIANG L, et al. LncRNA-Six1 is a target of miR-1611 that functions as a ceRNA to regulate Six1 protein expression and fiber type switching in chicken myogenesis[J]. Cells, 2018, 7(12). doi: 10.3390/cells7120243.
[58] DANAN M, SCHWARTZ S, EDELHEIT S, et al. Transcriptome-wide discovery of circular RNAs in archaea[J]. Nucleic Acids Res, 2012, 40(7): 3131-3142. doi: 10.1093/nar/gkr1009
[59] CHEN L L, YANG L. Regulation of circRNA biogenesis[J]. RNA Biol, 2015, 12(4): 381-388. doi: 10.1080/15476286.2015.1020271
[60] ENUKA Y, LAURIOLA M, FELDMAN M E, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor[J]. Nucleic Acids Res, 2016, 44(3): 1370-1383. doi: 10.1093/nar/gkv1367
[61] WU Q, WANG Y, CAO M, et al. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm[J]. Proc Natl Acad Sci USA, 2012, 109(10): 3938-3943. doi: 10.1073/pnas.1117815109
[62] LEGNINI I, DI TIMOTEO G, ROSSI F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis[J]. Mol Cell, 2017, 66(1): 22-37. doi: 10.1016/j.molcel.2017.02.017
[63] HANSEN T B, JENSEN T I, CLAUSEN B H, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495(7441): 384-388. doi: 10.1038/nature11993
[64] MEMCZAK S, JENS M, ELEFSINIOTI A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495(7441): 333-338. doi: 10.1038/nature11928
[65] DU W W, YANG W, LIU E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2[J]. Nucleic Acids Res, 2016, 44(6): 2846-2858. doi: 10.1093/nar/gkw027
[66] OUYANG H, CHEN X, LI W, et al. Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken[J]. Front Genet, 2018, 9. doi: 10.3389/fgene.2018.00172.
[67] CHEN X, OUYANG H, WANG Z, et al. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p[J]. Cells, 2018, 7(11). doi: 10.3390/cells7110199.
[68] CHEN B, YU J, GUO L, et al. Circular RNA circHIPK3 promotes the proliferation and differentiation of chicken myoblast cells by sponging miR-30a-3p[J]. Cells, 2019, 8(2). doi: 10.3390/cells8020177.
-
期刊类型引用(2)
1. 赵志刚,郭晓芹,赵传芳,吕文雪,于小亚,吕爽. 环介导等温扩增技术在动物细小病毒检测中的应用研究进展. 特产研究. 2025(02): 194-199 .  百度学术
百度学术
2. 陈文静,董章勇,宋汉达,罗梅. 环介导等温扩增技术在植物病原物检测中的应用. 仲恺农业工程学院学报. 2024(04): 48-54 .  百度学术
百度学术
其他类型引用(2)




 下载:
下载: