Resistance to blue ear disease and production performance assessment of CD163 gene-edited Large White pigs
-
摘要:目的
运用CRISPR/Cas9基因编辑技术和体细胞核移植技术获得CD163基因全敲除(CD163 gene knockout,CD163-KO)的克隆猪,验证该基因敲除大白猪的抗蓝耳病特性,并研究其与野生型大白猪在生理、生产和繁殖性能等方面的差别,评估CD163-KO大白猪的主要生产性能。
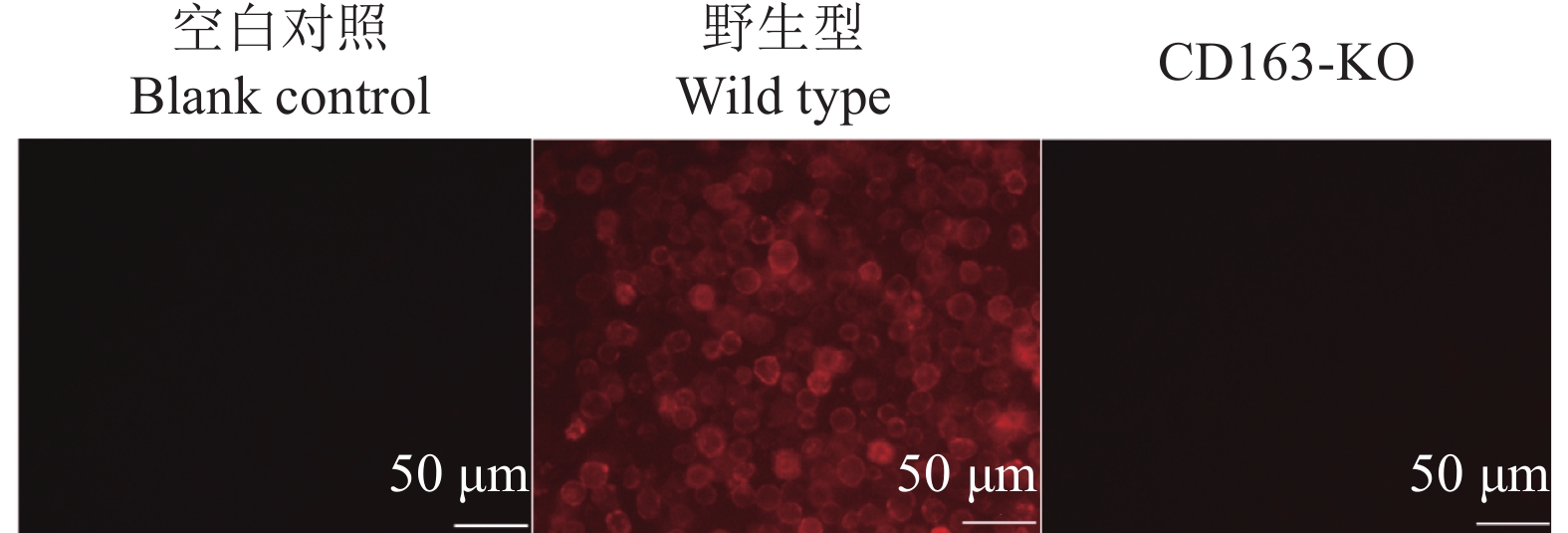
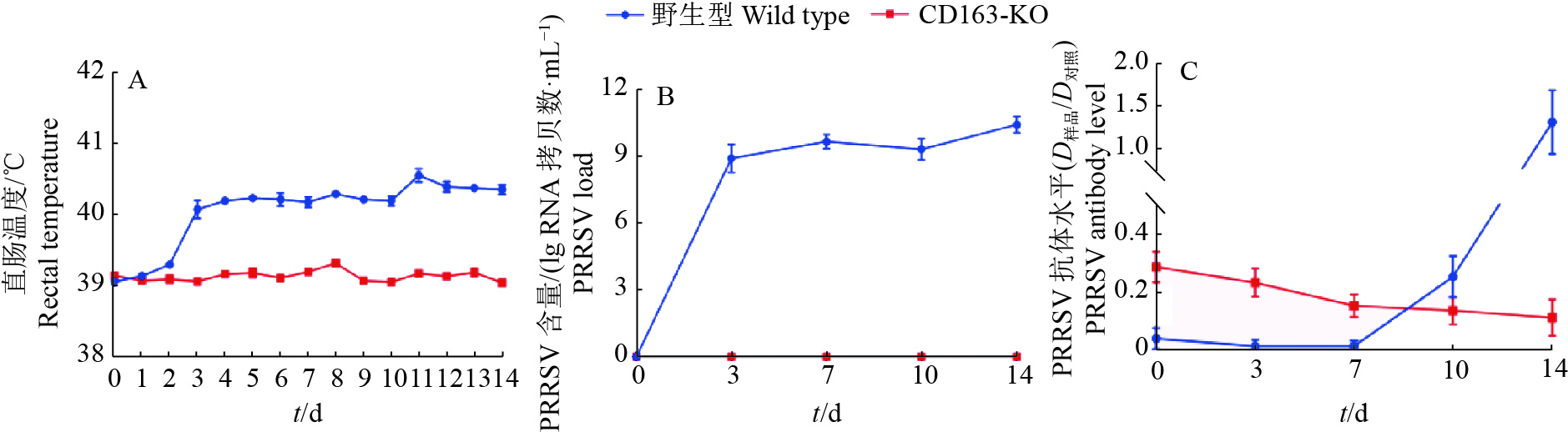
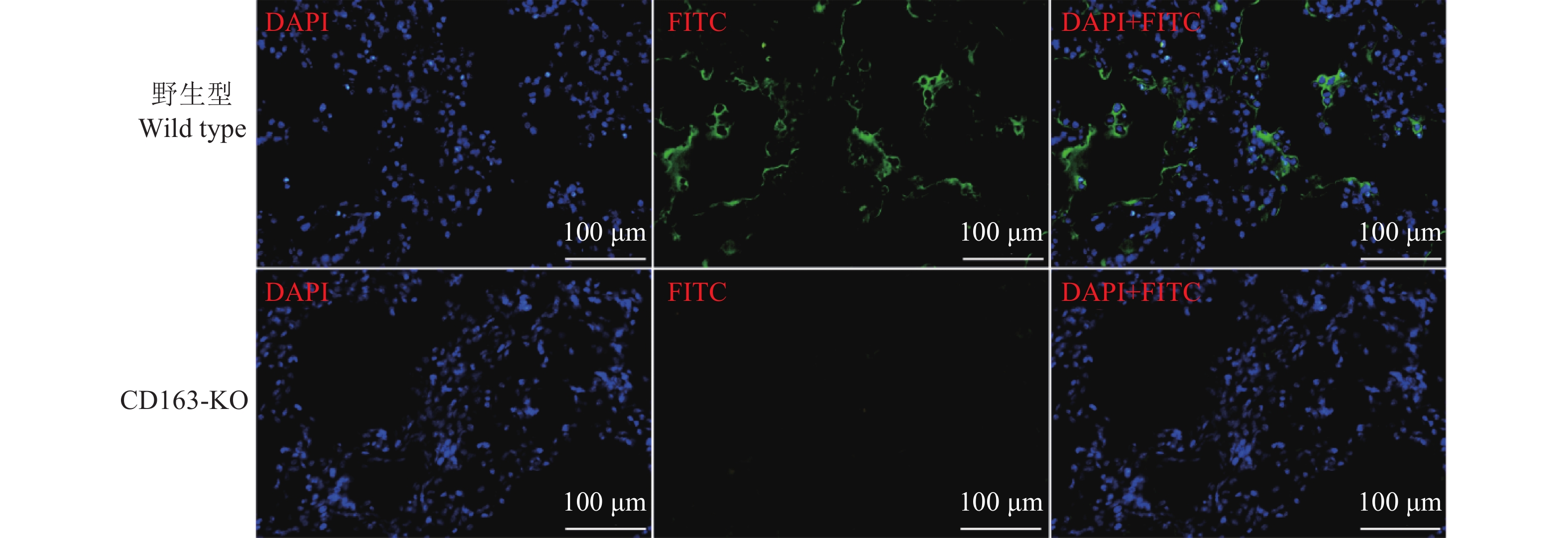
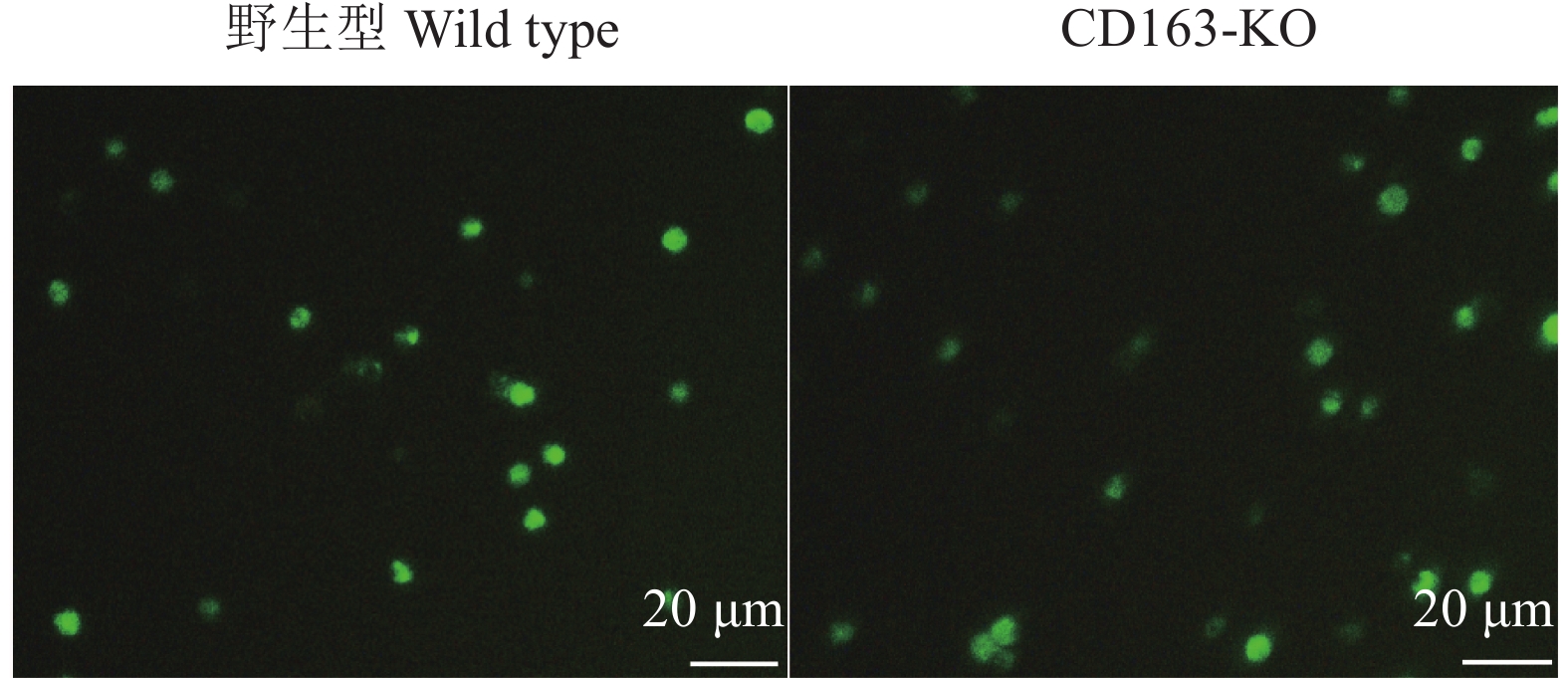
方法用猪繁殖与呼吸综合征病毒(Porcine reproductive and respiratory syndrome virus,PRRSV)流行毒株NADC30-like对本研究所获得的11头CD163-KO大白猪和5头年龄、体质量相当的野生型大白猪进行攻毒试验。连续14 d观察猪的临床症状,记录直肠温度变化,检测血清中PRRSV病毒含量和抗体水平。14 d后取肺部组织进行切片,通过PRRSV免疫荧光试验观察肺脏感染情况。采集CD163-KO和野生型大白猪肺泡巨噬细胞进行免疫染色,观察肺泡巨噬细胞表面CD163蛋白的表达情况;采集猪外周血单核细胞进行诱导分化,观察CD163-KO与野生型大白猪诱导分化的巨噬细胞对血红蛋白−结合珠蛋白复合物的摄取情况。最后,统计分析CD163-KO与野生型大白猪的生产性能及公猪的繁殖性能。
结果本研究获得的CD163-KO大白猪对蓝耳病NADC30-like毒株具有完全抗性,敲除CD163基因不影响巨噬细胞的生理功能,CD163-KO与野生型大白猪的生长性能和繁殖性能无明显差异。
结论本研究是对CD163-KO猪抵抗蓝耳病的又一佐证和补充,证明CD163基因敲除操作对生产性能没有潜在的负面影响,为抗蓝耳病猪的生物安全提供了支撑。
Abstract:ObjectiveThe purpose of this study was to generate CD163 gene knockout (CD163-KO) Large White pigs by CRISPR/Cas9 gene editing and somatic cell nuclear transfer technologies, investigate the resistance to blue ear disease and the biosafety effect including physiology, productive and reproductive performances of the gene knockout pigs, and assess the main production performances of CD163-KO Large White pigs.
MethodIn this study, the 11 CD163-KO pigs and five age- and body weight-matched wild type Large White pigs were challenged with NADC30-like strain of porcine reproductive and respiratory syndrome virus (PRRSV). The rectal temperature, PRRSV antibody and virus variation were monitored continuously for 14 days. The lung tissues were examined by immunofluorescence of PRRSV antigen. Expression of CD163 protein on the surface of pulmonary alveolar macrophages in wild type and CD163-KO Large White pigs were examined through immunofluorescence staining. We compared the differentiation potential of monocytes into macrophages between CD163-KO and wild type pigs, and observed their uptake capacities to hemoglobin-haptoglobin complex. In addition, we analyzed the growth and reproductive production of the boars between CD163-KO pigs and wild type control to assess their biosafety and breeding value.
ResultCD163-KO pigs were completely resistant to NADC30-like strain without impairing the biological function associated with the modified gene, as well as productive and reproductive performances.
ConclusionThis study is an evidence and supplement of CD163-KO pigs resistance to blue ear disease, and demonstrates that CD163 gene knockout has no potentially negative effects on production performance, which provides evidences for the biosecurity of CD163-KO pigs.
-
-
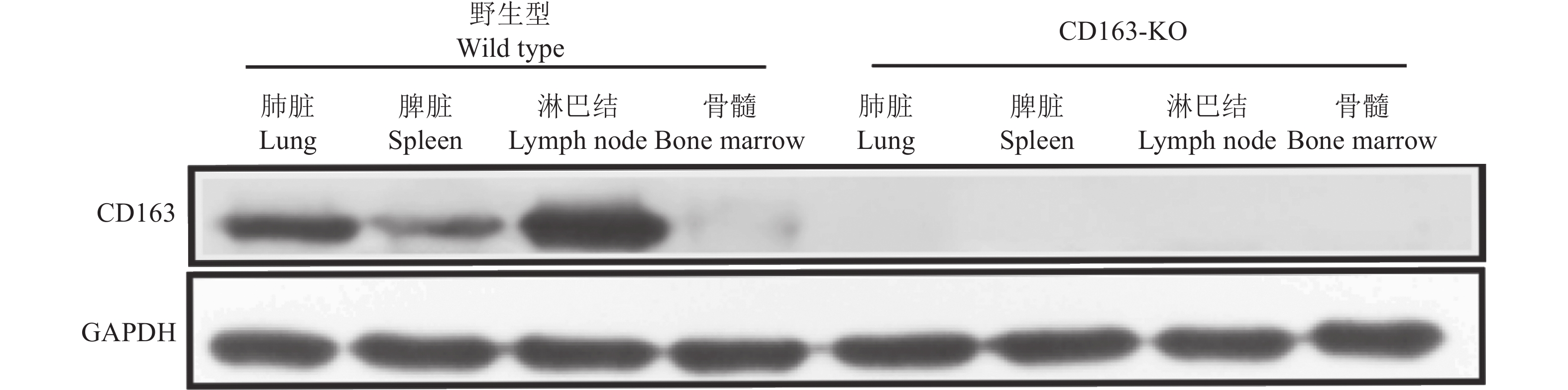
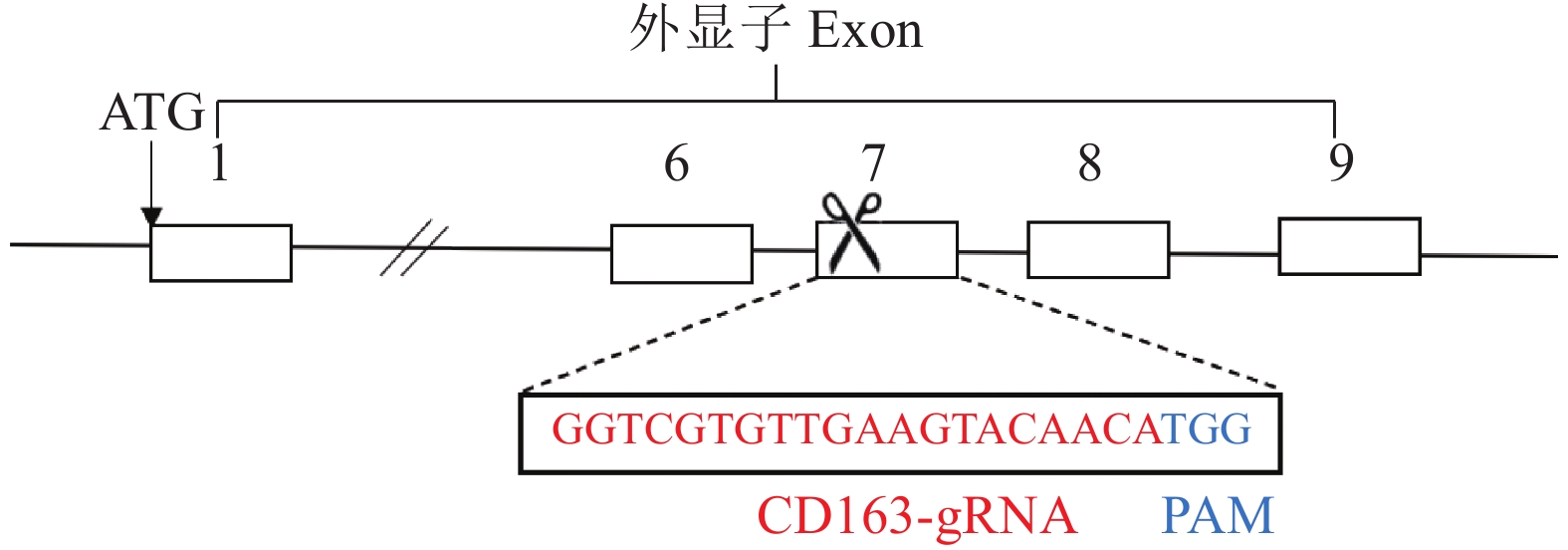
表 1 阳性单克隆细胞及CD163-KO大白猪测序结果
Table 1 Sequencing results of positive monoclonal cells and CD163-KO Large White pigs
类型 Type 靶位点序列(5′→3′) Sequence of target site 插入/缺失 Insertion/ Deletion 数量 Number 野生型大白猪 Wild type Large White pig GGTCGTGTTGAAGTACAACATGG 阳性单克隆细胞 Positive monoclonal cell GGTCGTGTTGAAGTACAAACATGG +A 3 CD163-KO大白猪 CD163-KO Large White pig GGTCGTGTTGAAGTACAAACATGG +A 18 表 2 野生型(n=10)和CD16-KO (n=5)大白猪生产性能比较1)
Table 2 Comparison of the productive performance of wild type and CD163-KO Large White pigs
项目 Item 野生型 Wild type CD163-KO P 出生体质量/kg Birth body weight 2.04±0.13 1.80±0.15 0.277 115 kg体长/cm Body length of 115 kg 119.8±1.2 119.2±1.0 0.746 115 kg体高/cm Body height of 115 kg 68.6±0.6 68.4±0.8 0.841 115 kg背膘厚/mm Backfat thickness of 115 kg 12.069±0.401 11.702±0.495 0.592 115 kg眼肌面积/cm2Eye muscle area of 115 kg 35.670±1.202 36.320±1.090 0.736 30 ~115 kg平均日增体质量/kg Average daily gain of body weight during 30−115 kg 1.075±0.047 1.008±0.068 0.428 30~115 kg饲料转化率 Feed/gain ratio during 30−115 kg 2.326±0.085 2.548±0.154 0.191 1)表中数据为平均值±标准误,P>0.05表示两组间差异不显著 (t检验) 1) Data are expressed as the mean ± standard error, P > 0.05 means there is no significant difference between two sets ( t test) 表 3 野生型(n=20)与CD163-KO (n=10)大白猪精液质量比较1)
Table 3 Comparison of the quality of the semen from wild type and CD163-KO Large White pigs
项目 Item 野生型 Wild type CD163-KO P 精子总数/亿 Number of sperms 680.643 ± 84.535 685.726 ± 99.930 0.971 1∶1稀释后精子活力 Sperm motility after 1∶1 dilution 0.675 ± 0.013 0.700 ± 0.000 0.219 1∶1稀释后精子密度/(亿·mL−1) Sperm density after 1∶1 dilution 1.685 ± 0.195 1.578 ± 0.239 0.747 精子畸形率/% Abnormal rate of sperm 11.092 ± 1.358 10.000 ± 0.964 0.606 1)表中数据为平均值±标准误,P>0.05表示两组间没有显著差异(t检验) 1) Data are expressed as the mean ± standard error, P > 0.05 means there is no significant difference between two sets ( ttest) -
[1] HAN J, ZHOU L, GE X, et al. Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus[J]. Veterinary Microbiology, 2017, 209: 30-47. doi: 10.1016/j.vetmic.2017.02.020
[2] TIAN K, YU X, ZHAO T, et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark[J]. PLoS One, 2007, 2(6): e526. doi: 10.1371/journal.pone.0000526
[3] ZHAO K, YE C, CHANG X, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China[J]. Journal of Virology, 2015, 89(20): 10712-10716. doi: 10.1128/JVI.01446-15
[4] WANG H M, LIU Y G, TANG Y D, et al. A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains[J]. Transboundary and Emerging Diseases, 2018, 65(4): 1078-1086. doi: 10.1111/tbed.12852
[5] YU Y, ZHANG Q, CAO Z, et al. Recent advances in porcine reproductive and respiratory syndrome virus NADC30-like research in China: Molecular characterization, pathogenicity, and control[J]. Frontiers in Microbiology, 2022, 12: 791313. doi: 10.3389/fmicb.2021.791313.
[6] ZHOU L, YANG B, XU L, et al. Efficacy evaluation of three modified-live virus vaccines against a strain of porcine reproductive and respiratory syndrome virus NADC30-like[J]. Veterinary Microbiology, 2017, 207: 108-116. doi: 10.1016/j.vetmic.2017.05.031
[7] CHAI W, LIU Z, SUN Z, et al. Efficacy of two porcine reproductive and respiratory syndrome (PRRS) modified-live virus (MLV) vaccines against heterologous NADC30-like PRRS virus challenge[J]. Veterinary Microbiology, 2020, 248: 108805. doi: 10.1016/j.vetmic.2020.108805.
[8] RENUKARADHYA G J, MENG X J, CALVERT J G, et al. Inactivated and subunit vaccines against porcine reproductive and respiratory syndrome: Current status and future direction[J]. Vaccine, 2015, 33(27): 3065-3072. doi: 10.1016/j.vaccine.2015.04.102
[9] 熊胜利, 龙清孟, 陈大方, 等. 抗病育种技术在引进美国SPF种猪后代选育中应用探究[J]. 养猪, 2014(3): 73-74. doi: 10.13257/j.cnki.21-1104/s.2014.03.028 [10] YUAN H, YANG L, ZHANG Y, et al. Current status of genetically modified pigs that are resistant to virus infection[J]. Viruses, 2022, 14(2): 417. doi: 10.3390/v14020417.
[11] PROUDFOOT C, LILLICO S, TAIT-BURKARD C. Genome editing for disease resistance in pigs and chickens[J]. Animal Frontiers, 2019, 9(3): 6-12. doi: 10.1093/af/vfz013
[12] WHITWORTH K M, ROWLAND R R R, EWEN C L, et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus[J]. Nature Biotechnology, 2016, 34(1): 20-22. doi: 10.1038/nbt.3434
[13] YANG H, ZHANG J, ZHANG X, et al. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus[J]. Antiviral Research, 2018, 151: 63-70.
[14] BURKARD C, LILLICO S G, REID E, et al. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function[J]. PLOS Pathogens, 2017, 13(2): e1006206. doi: 10.1371/journal.ppat.1006206
[15] WELCH S K W, CALVERT J G. A brief review of CD163 and its role in PRRSV infection[J]. Virus Research, 2010, 154(1/2): 98-103. doi: 10.1016/j.virusres.2010.07.018
[16] XU Y, WU S, LI Y, et al. A porcine alveolar macrophage cell line stably expressing CD163 demonstrates virus replication and cytokine secretion characteristics similar to primary alveolar macrophages following PRRSV infection[J]. Veterinary Microbiology, 2020, 244: 108690. doi: 10.1016/j.vetmic.2020.108690.
[17] XU K, ZHOU Y, MU Y, et al. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance[J]. eLife, 2020, 9: e57132.
[18] 韩晓松, 高杨, 刘海龙, 等. 利用CRISPR/Cas9技术制备CD163基因SRCR5序列敲除猪[J]. 农业生物技术学报, 2020, 28(9): 1535-1542. [19] GUO C, WANG M, ZHU Z, et al. Highly efficient generation of pigs harboring a partial deletion of the CD163 SRCR5 domain, which are fully resistant to porcine reproductive and respiratory syndrome virus 2 infection[J]. Frontiers in Immunology, 2019, 10: 1846. doi: 10.3389/fimmu.2019.01846.
[20] CHEN J, WANG H, BAI J, et al. Generation of pigs resistant to highly pathogenic-porcine reproductive and respiratory syndrome virus through gene editing of CD163[J]. International Journal of Biological Sciences, 2019, 15(2): 481-492. doi: 10.7150/ijbs.25862
[21] VAN GORP H, VAN BREEDAM W, DELPUTTE P L, et al. Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus[J]. Journal of General Virology, 2008, 89: 2943-2953. doi: 10.1099/vir.0.2008/005009-0
[22] ZHANG Q, YOO D. PRRS virus receptors and their role for pathogenesis[J]. Veterinary Microbiology, 2015, 177(3/4): 229-241.
[23] VAN BREEDAM W, VERBEECK M, CHRISTIAENS I, et al. Porcine, murine and human sialoadhesin (Sn/Siglec-1/CD169): Portals for porcine reproductive and respiratory syndrome virus entry into target cells[J]. Journal of General Virology, 2013, 94: 1955-1960.
[24] GRAVERSEN J H, MADSEN M, MOESTRUP S K. CD163: A signal receptor scavenging haptoglobin-hemoglobin complexes from plasma[J]. International Journal of Biochemistry & Cell Biology, 2002, 34(4): 309-314.
[25] KRISTIANSEN M, GRAVERSEN J H, JACOBSEN C, et al. Identification of the haemoglobin scavenger receptor[J]. Nature, 2001, 409(6817): 198-201. doi: 10.1038/35051594
[26] SCHAER C A, VALLELIAN F, IMHOF A, et al. CD163-expressing monocytes constitute an endotoxin-sensitive Hb clearance compartment within the vascular system[J]. Journal of Leukocyte Biology, 2007, 82(1): 106-110. doi: 10.1189/jlb.0706453
[27] 石俊松, 周荣, 曾海玉, 等. 体细胞克隆猪繁殖性能及后代生长性能评估[J]. 华南农业大学学报, 2019, 40(S1): 100-103. [28] ADACHI N, YAMAGUCHI D, WATANABE A, et al. Growth, reproductive performance, carcass characteristics and meat quality in F1 and F2 progenies of somatic cell-cloned pigs[J]. Joural of Reproduction and Development, 2014, 60(2): 100-105. doi: 10.1262/jrd.2012-167
[29] KAWARASAKI T, ENYA S, OTAKE M, et al. Reproductive performance and expression of imprinted genes in somatic cell cloned boars[J]. Animal Science Journal, 2017, 88(11): 1801-1810. doi: 10.1111/asj.12838



 下载:
下载: