Development of an automatic system for SNP detection in diploid fragment sequencing
-
摘要:目的
开发基于模式识别方法的二倍体片段测序中单核苷酸多态性(Single nucleotide polymorphism,SNP)自动检测系统,提高检测的准确性。
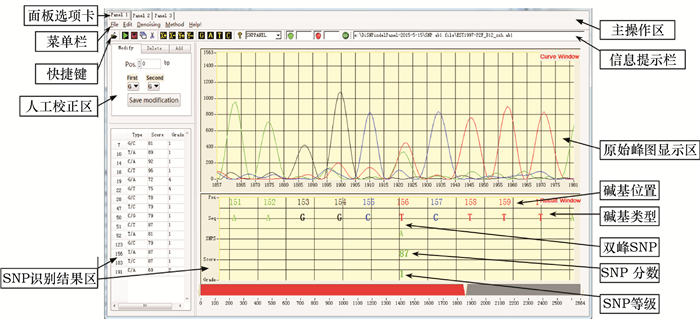
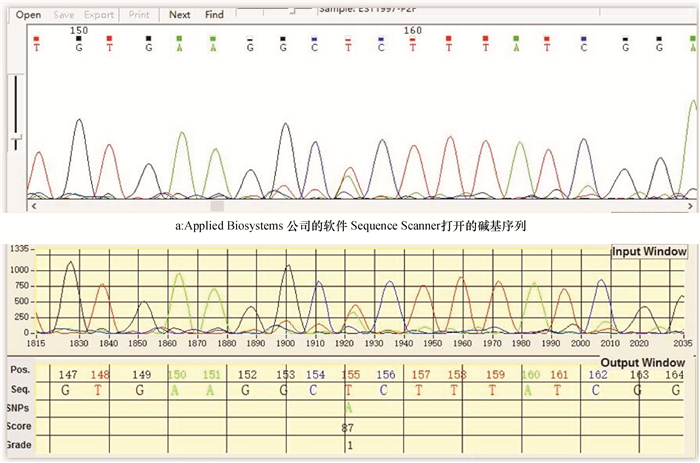
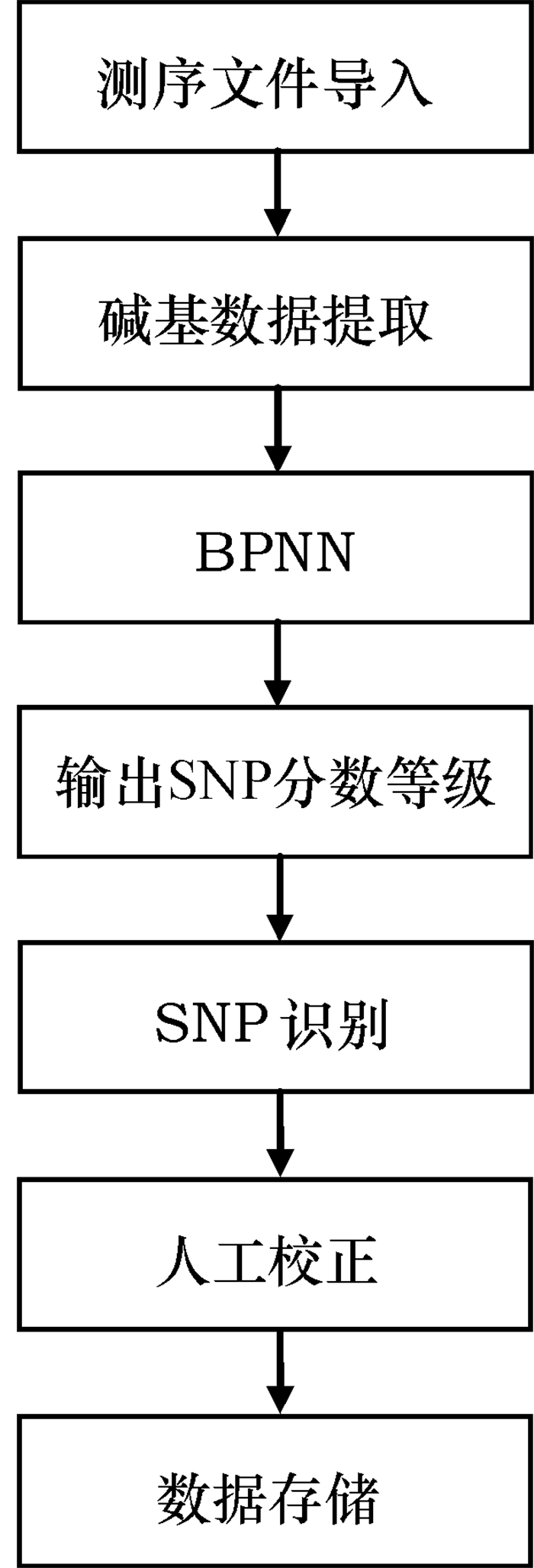
方法采用LabWindows/CVI 9.0开发平台,结合Matlab函数库编程,以二倍体PCR片段测序的.ab1或.scf格式文件作为源数据,首先分离出碱基G、A、T和C,进行一维离散小波滤波,再对各碱基处的波形进行典型特征提取,最后运用基于反向传播神经网络的分类器完成SNP识别和判断。
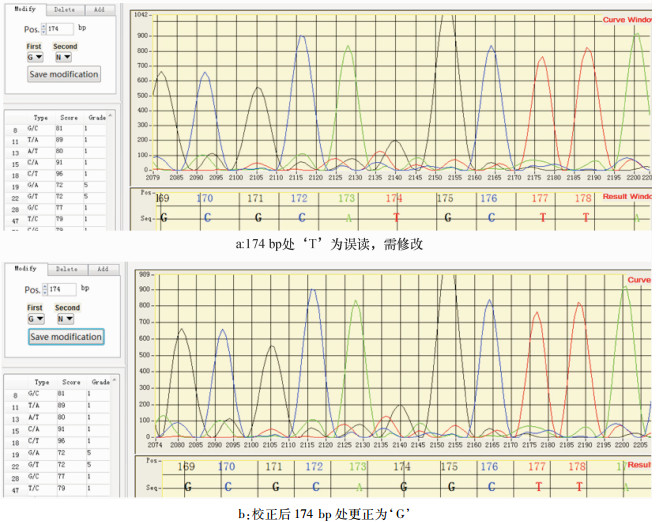
结果系统界面友好、运行稳定。SNP等级分为6级,允许用户对可疑的SNP进行人工修正,对尾叶桉Eucalyptus urophylla的26个测序序列143个SNP的测试中检测准确率、假阳性率和假阴性率均明显优于之前的类似软件。
结论本文所构建的SNP自动检测系统准确性高,不需参考序列,可用于二倍体PCR片段测序中SNP的高效检测。
Abstract:ObjectiveThis study aims to develop a pattern-recognition based system for automatic single nucleotide polymorphism (SNP)detection in diploid fragment sequencing and improve the detection accuracy.
MethodThe LabWindows/CVI 9.0 platform and Matlab environment were combined for analyzing.ab1 or.scf files generated in diploid PCR fragment sequencing. Firstly, four bases G, A, T and C were separated for eliminating noise through one-dimensional discrete wavelet filtering, following with extraction of typical features of each base position (peak) from a fluorescence curve. A classifier based on back-propagation neural network was then used for SNP recognition and diagnosis.
ResultThis established system was characterized by friendly interface, stable operation and manual modification accessibility. It classified the SNP reliability into six grades. Performance test with 143 SNPs of 26 sequencing fragments from Eucalyptus urophylla demonstrated that our system outperformed three previously reported software packages in detecting accuracy, false positive and false negative rates.
ConclusionOur system has a high rate of accuracy without the need for a reference sequence. It could be used for efficient SNP detection in diploid PCR fragment sequencing.
-
Keywords:
- diploid /
- sequencing /
- SNP detection /
- pattern recognition /
- Eucalyptus urophylla
-
表 1 不同软件识别SNP的结果对比1)
Table 1 Comparison of software performance in SNP detection

-
[1] OSSOWSKI S, SCHNEEBERGER K, CLARK R M, et al. Sequencing of natural strains of Arabidopsis thaliana wit short reads[J]. Genome Res, 2008, 18(12): 2024-2033. doi: 10.1101/gr.080200.108
[2] 唐立琼, 肖层林, 王伟平. SNP分子标记的研究及其应用进展[J].中国农学通报, 2012, 28(12): 154-158. http://d.old.wanfangdata.com.cn/Periodical/zgnxtb201212028 [3] 许家磊, 王宇, 后猛, 等. SNP检测方法的研究进展[J].分子植物育种, 2015, 13(2): 475-482. http://d.old.wanfangdata.com.cn/Periodical/yc200601021 [4] WECKX S, DEL-FAVERO J, RADEMAKERS R, et al. novoSNP, a novel computational tool for sequence variation discovery[J]. Genome Res, 2005, 15(3): 436-442. doi: 10.1101/gr.2754005
[5] MATTHEW S, JAMES S, ROBERTSON P D, et al. Automating sequence-based detection and genotyping of single nucleotide polymorphisms (SNPs) from diploid samples[J]. Nat Genet, 2006, 38(3): 375-381. doi: 10.1038/ng1746
[6] DENG J Z, HUANG H S, YU X L, et al. DiSNPindel: Improved intra-individual SNP and InDel detection in direct amplicon sequencing of a diploid[J]. BMC Bioinformatics, 2015, 16: 343. doi: 10.1186/s12859-015-0790-y
[7] 仇志平, 李树军. LabWindows/CVI虚拟仪器软件在测试领域中的应用[J].计算机工程与设计, 2007, 28(22): 5544-5548. doi: 10.3969/j.issn.1000-7024.2007.22.065 [8] 刘君华.虚拟程序编程语言LabWindows/CVI编程[M].北京:电子工业出版社, 2001. [9] 肖伟, 刘忠, 曾新勇, 等. MATLAB程序设计与应用[M].北京:清华大学出版社, 2005. [10] BUI T D, CHEN G. Translation-invariant denoising using multiwavelets[J]. IEEE Trans Sig Proc, 1998, 46(12): 3414-3420. doi: 10.1109/78.735315
[11] PAN Q, ZHANG P, DAI G, et al. Two denoising methods by wavelet transform[J]. IEEE Trans Sig Proc, 1999, 47(12): 3401-3406. doi: 10.1109/78.806084
[12] MCKEOWN J J, STELLA F, HALL G. Some numerical aspects of the training problem for feed-forward neural nets[J]. Neural Netw, 1997, 10(9): 1455-1463. doi: 10.1016-S0893-6080(97)00015-4/
[13] 黄华盛. 基于模式识别的二倍体个体内SNP和InDel自动检测[D]. 广州: 华南农业大学, 2014. [14] YU X, GUO Y, ZHANG X, et al. Integration of EST-CAPS markers into genetic maps of and Eucalgptus urophylla and E.tereticornis and their alignment with E. grandis genome sequence[J]. Silvae Genet, 2012, 61(6): 247-255.
[15] STUDER A, ZHAO Q, ROSS-IBARRA J, et al. Identification of a functional transposon insertion in the maize domestication gene tb1[J]. Nat Genet, 2011, 43(11): 1160-1163. doi: 10.1038/ng.942
[16] NGAMPHIW C, KULAWONGANUNCHAI S, ASSAWAMAKIN A, et al. VarDetect: A nucleotide sequence variation exploratory tool[J]. BMC Bioinformatics, 2008, 9(S12): 9. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_2638149



 下载:
下载: